Deep Learning Method Applied to Autonomous Image Diagnosis for Prick Test
Abstract
:1. Introduction
2. Methods
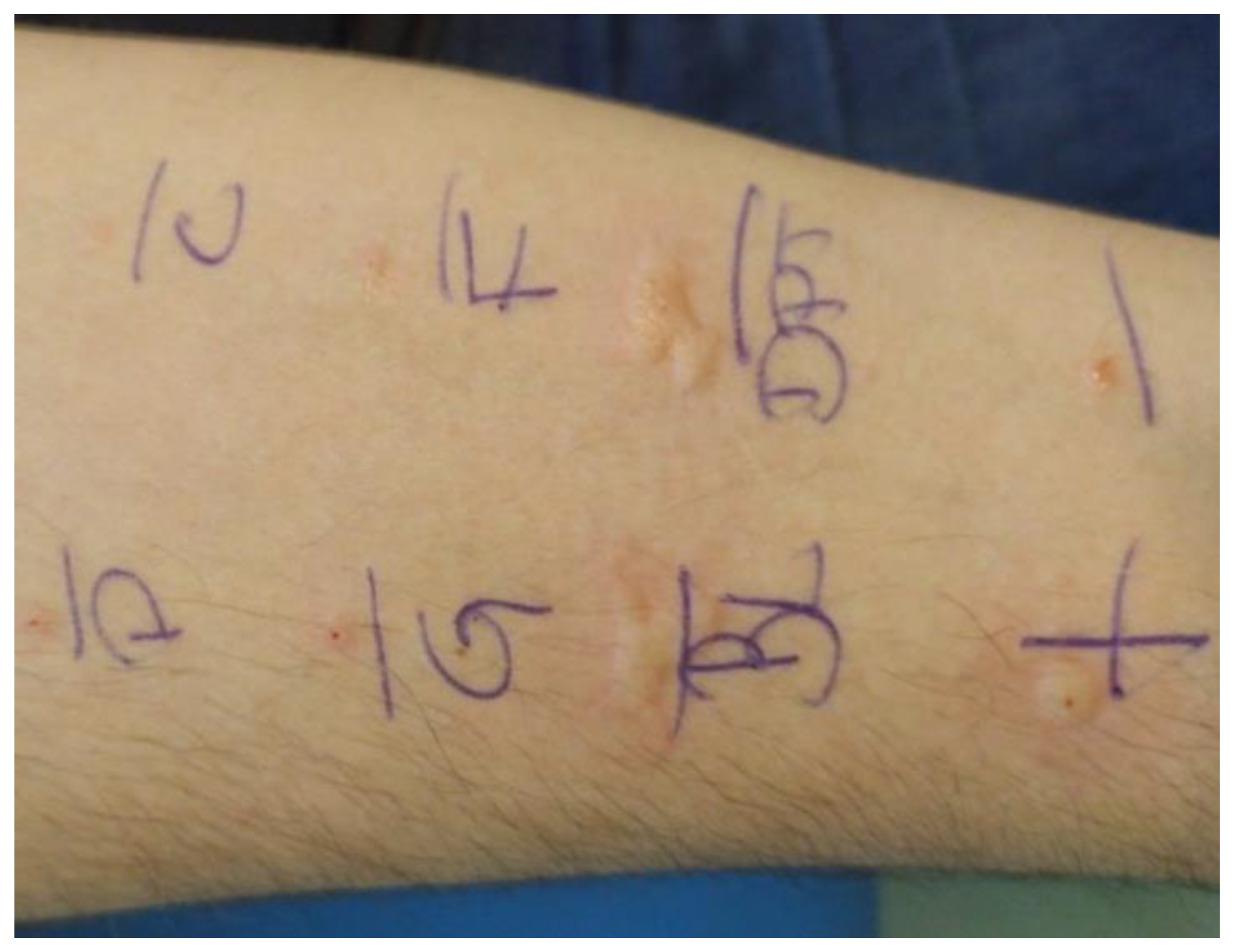
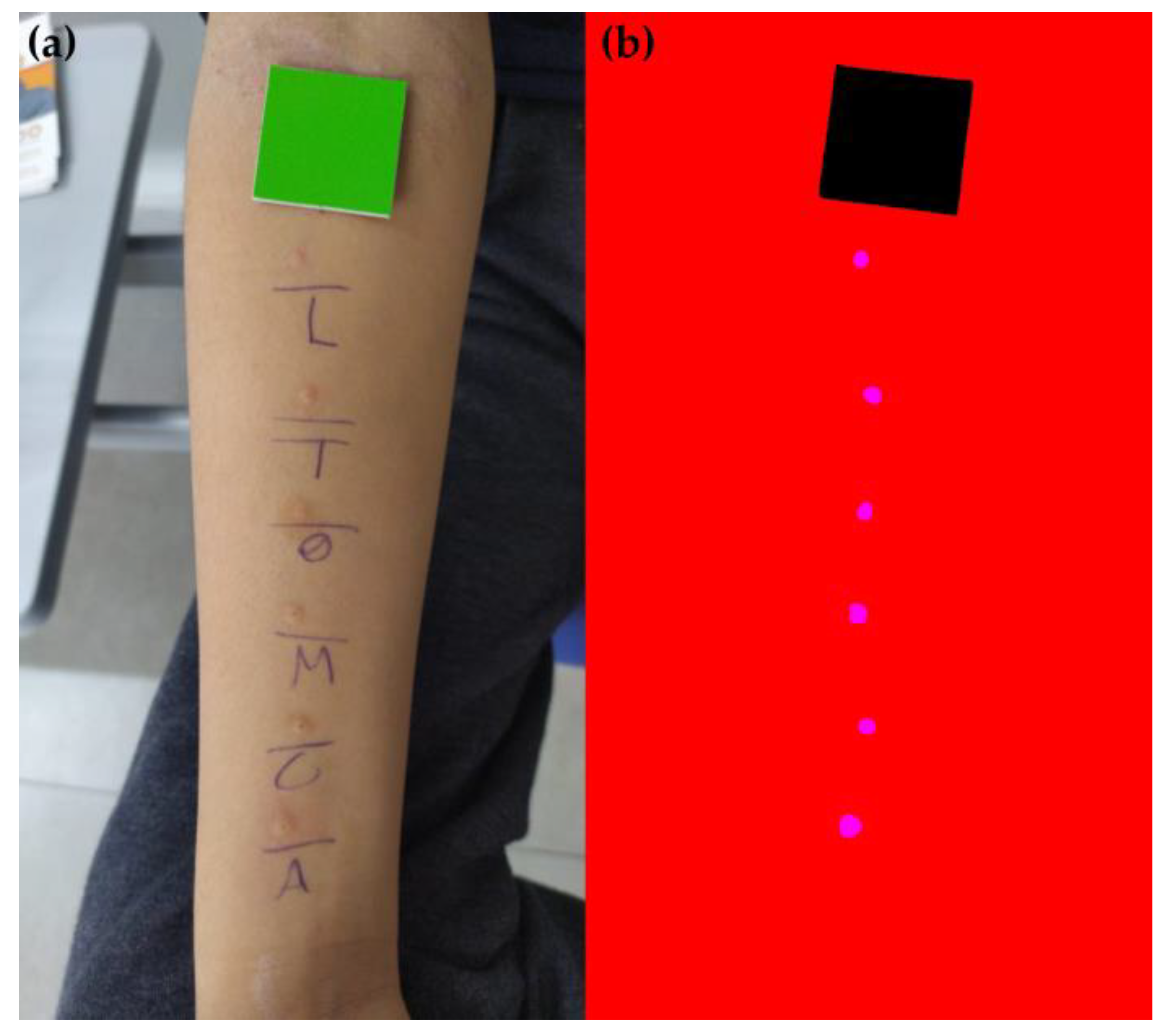
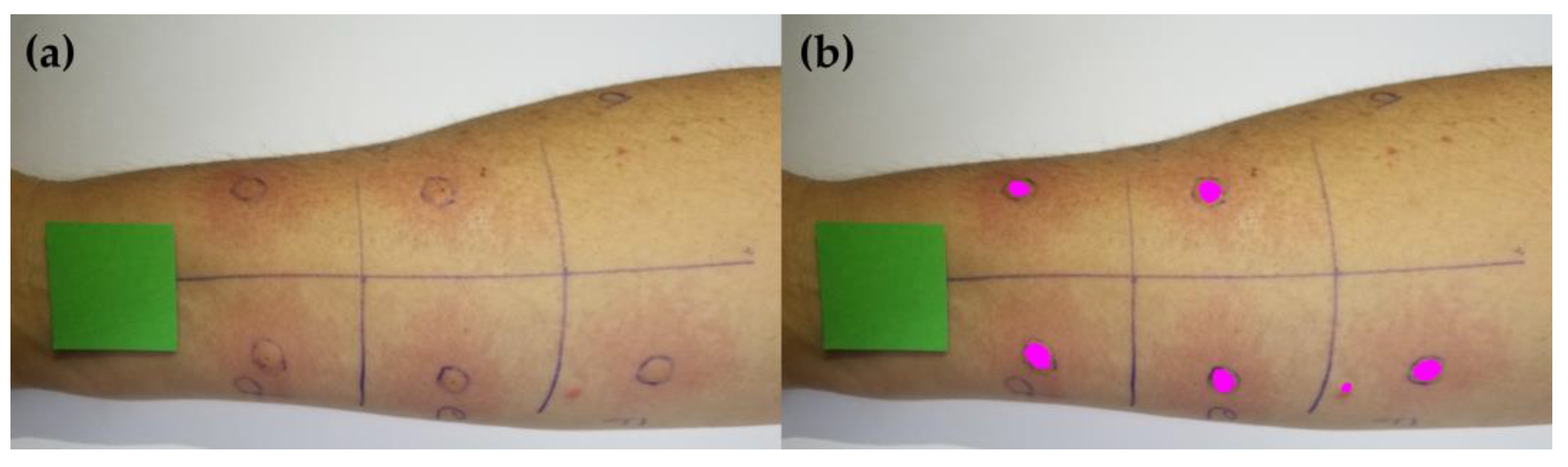
2.1. Acquisition of Skin Prick Test Photos
2.2. Dataset Standardization
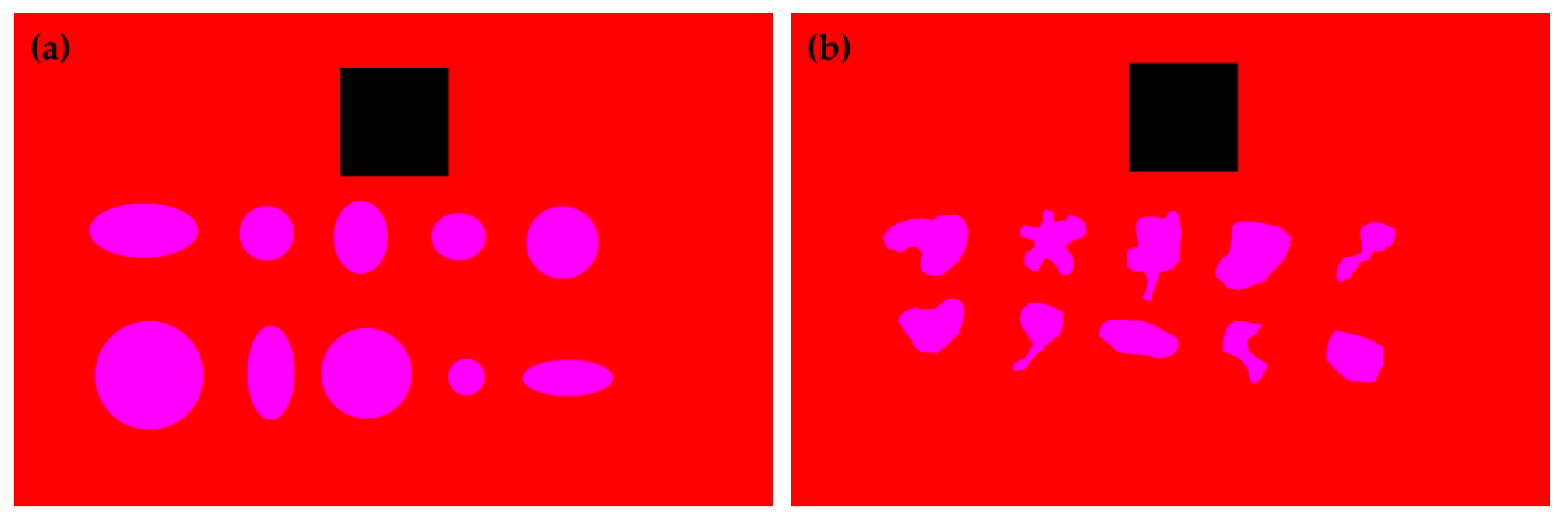
2.3. Machine Learning Model Training and Wheal Clustering
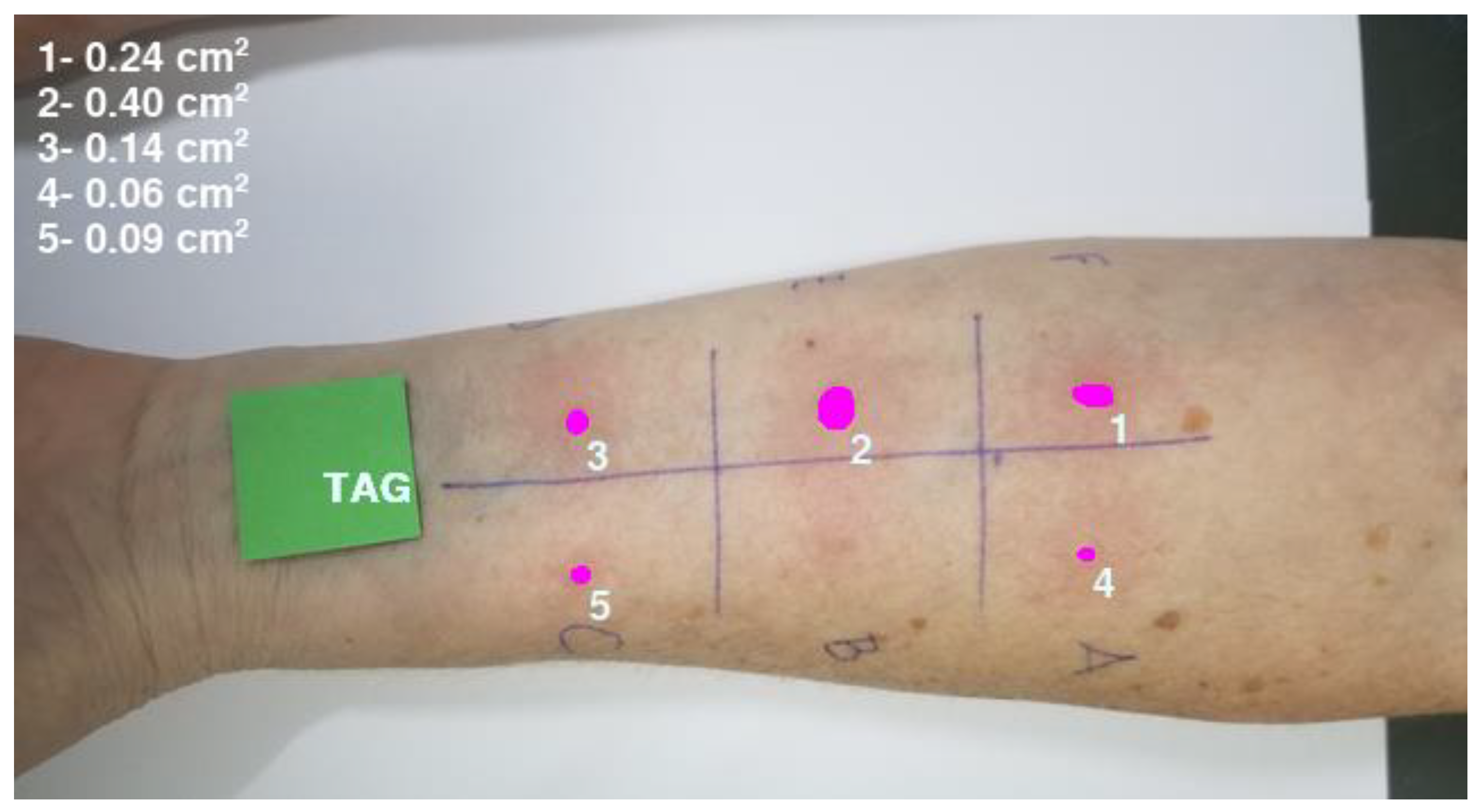
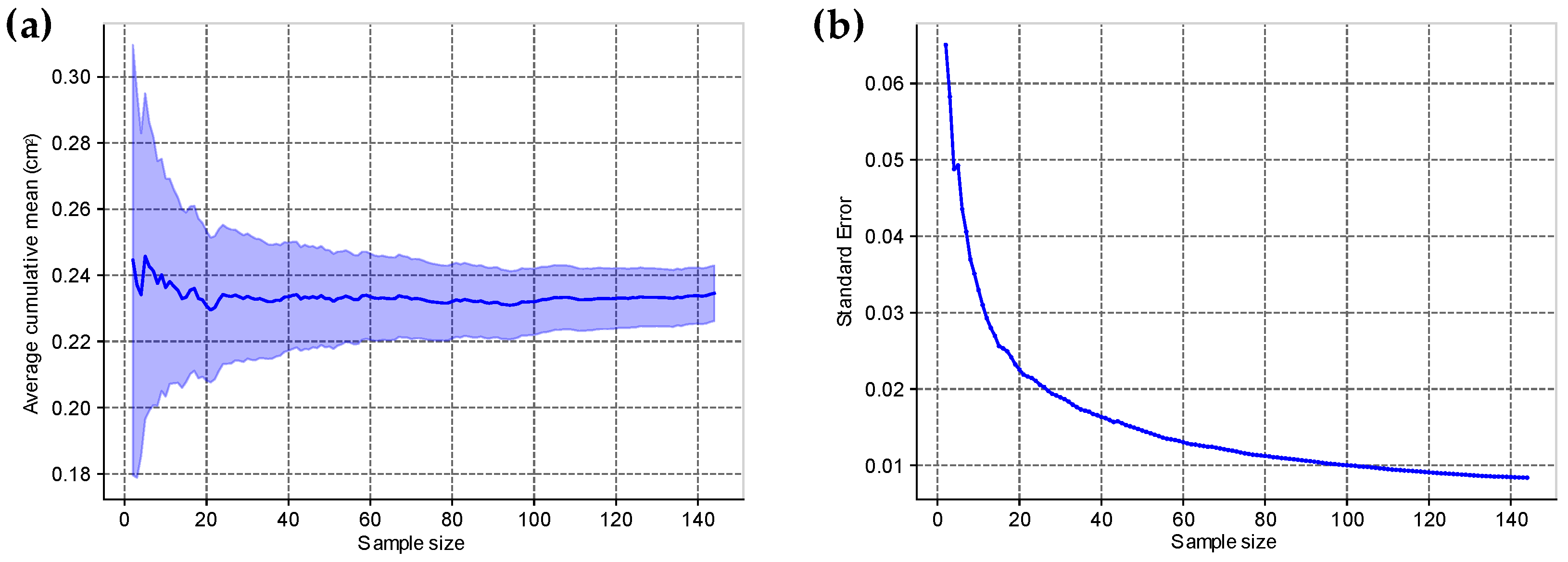
2.4. Evaluation of the Machine Learning Model Performance
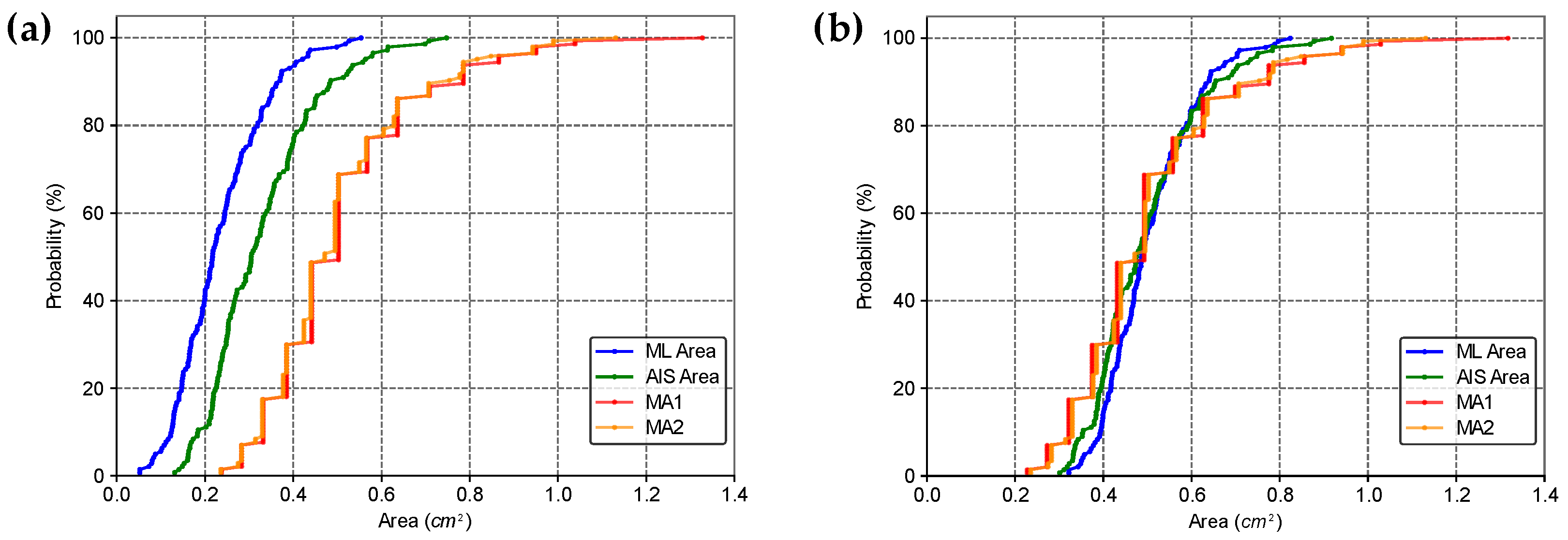
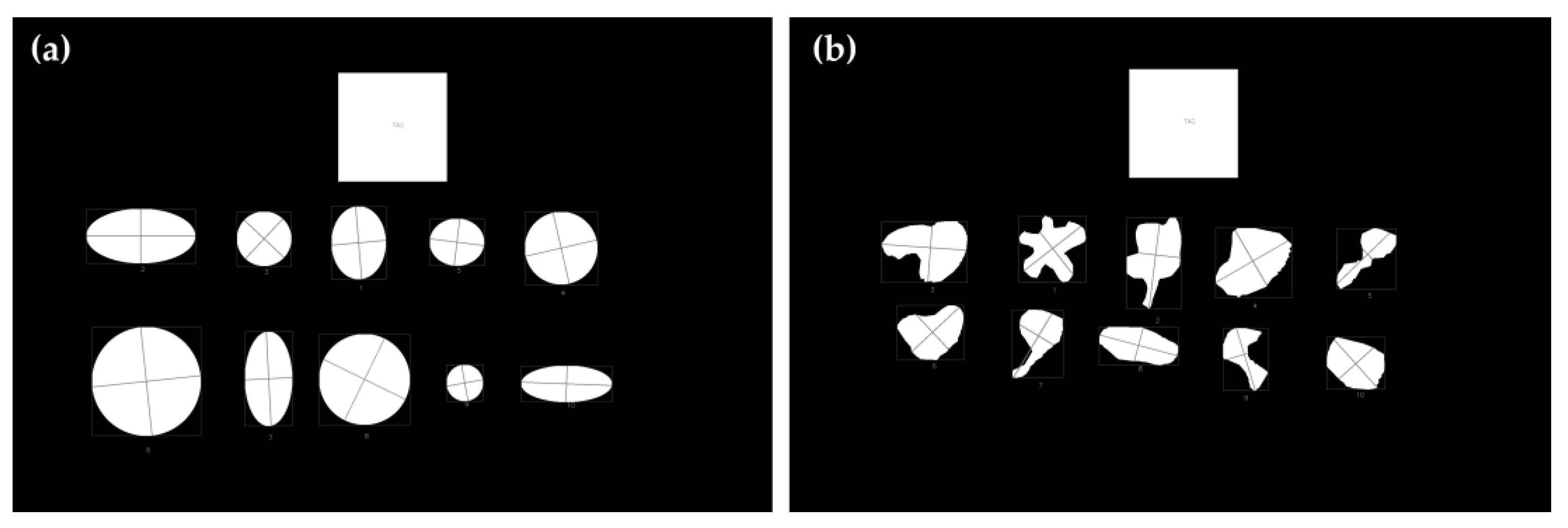
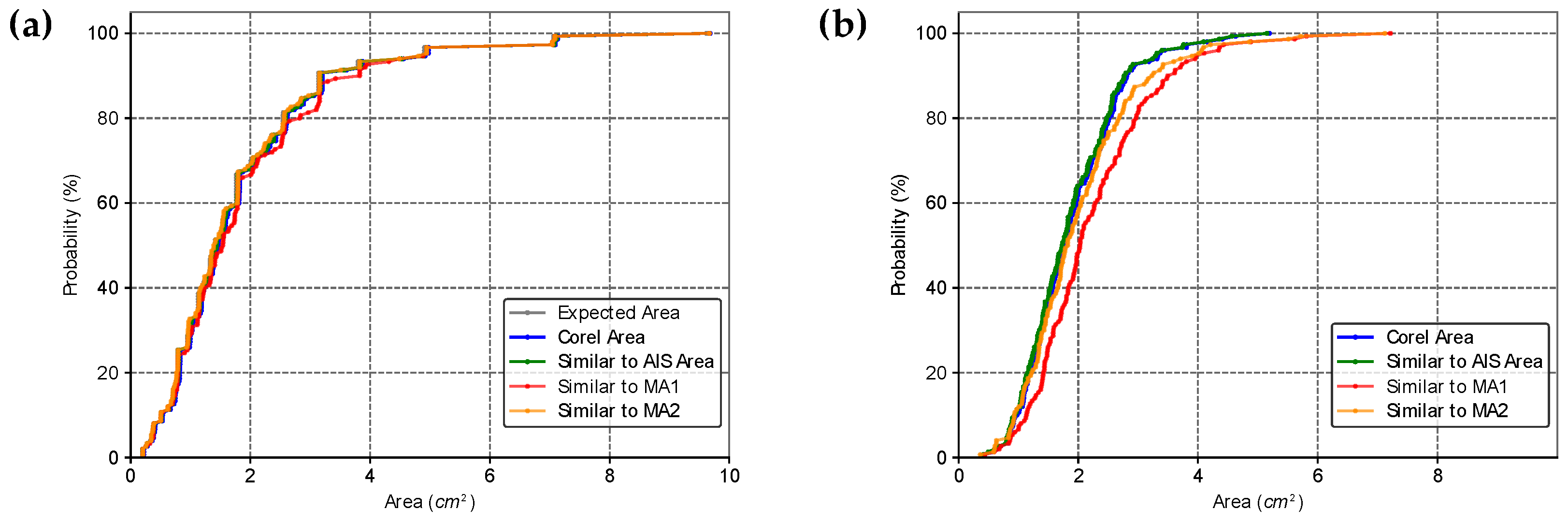
2.5. Comparison of Area Estimates for Regular and Irregular Geometric Shapes Using Different Methods
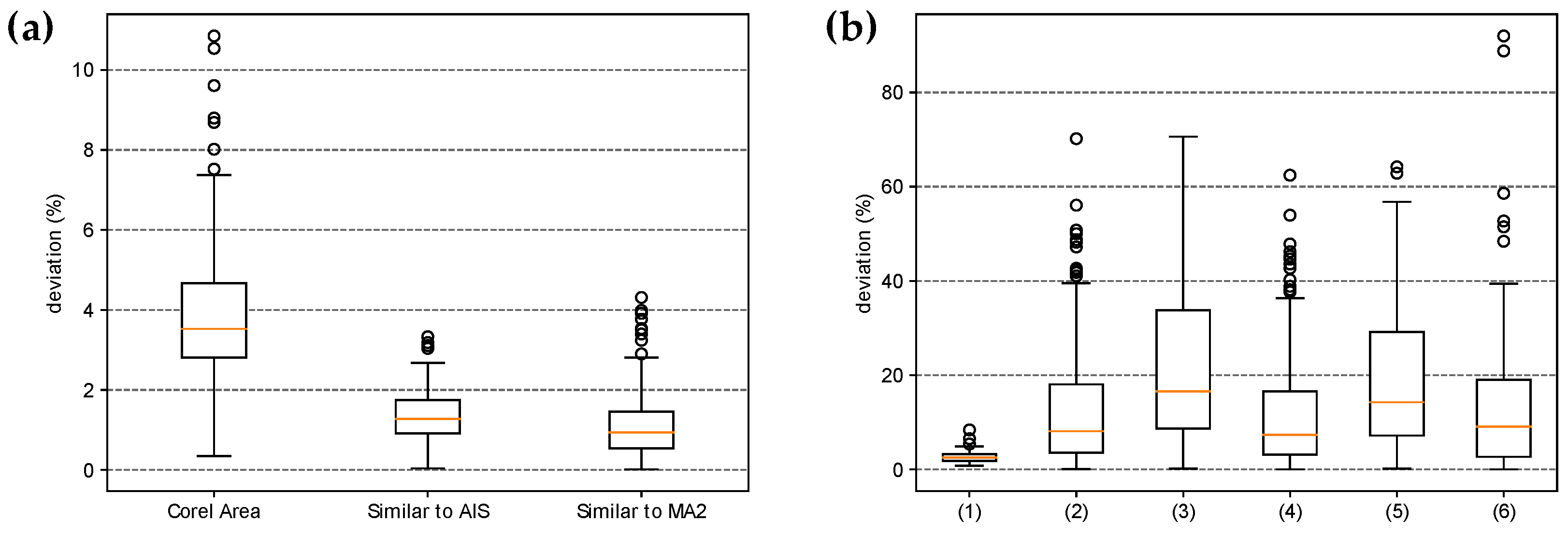
3. Results
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frati, F.; Incorvaia, C.; Cavaliere, C.; Di Cara, G.; Marcucci, F.; Esposito, S.; Masieri, S. The Skin Prick Test. J. Biol. Regul. Homeost. Agents 2018, 32, 19–24. [Google Scholar] [PubMed]
- Knight, V.; Wolf, M.L.; Trikha, A.; Curran-Everett, D.; Hiserote, M.; Harbeck, R.J. A Comparison of Specific IgE and Skin Prick Test Results to Common Environmental Allergens Using the HYTECTM 288. J. Immunol. Methods 2018, 462, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Topal, S.; Karaman, B.; Aksungur, V. Variables Affecting Interpretation of Skin Prick Test Results. Indian J. Dermatol. Venereol. Leprol. 2017, 83, 200. [Google Scholar] [CrossRef]
- Van Der Valk, J.P.M.; Gerth Van Wijk, R.; Hoorn, E.; Groenendijk, L.; Groenendijk, I.M.; De Jong, N.W. Measurement and Interpretation of Skin Prick Test Results. Clin. Transl. Allergy 2015, 6, 8. [Google Scholar] [CrossRef]
- Haahtela, T.; Burbach, G.J.; Bachert, C.; Bindslev-Jensen, C.; Bonini, S.; Bousquet, J.; Bousquet-Rouanet, L.; Bousquet, P.J.; Bresciani, M.; Bruno, A.; et al. Clinical Relevance Is Associated with Allergen-specific Wheal Size in Skin Prick Testing. Clin. Exp. Allergy 2014, 44, 407–416. [Google Scholar] [CrossRef]
- Heinzerling, L.; Mari, A.; Bergmann, K.-C.; Bresciani, M.; Burbach, G.; Darsow, U.; Durham, S.; Fokkens, W.; Gjomarkaj, M.; Haahtela, T.; et al. The Skin Prick Test—European Standards. Clin. Transl. Allergy 2013, 3, 3. [Google Scholar] [CrossRef]
- Justo, X.; Díaz, I.; Gil, J.J.; Gastaminza, G. Medical Device for Automated Prick Test Reading. IEEE J. Biomed. Health Inform. 2018, 22, 895–903. [Google Scholar] [CrossRef]
- Justo, X.; Díaz, I.; Gil, J.J.; Gastaminza, G. Prick Test: Evolution towards Automated Reading. Allergy 2016, 71, 1095–1102. [Google Scholar] [CrossRef]
- Andersen, H.H.; Lundgaard, A.C.; Petersen, A.S.; Hauberg, L.E.; Sharma, N.; Hansen, S.D.; Elberling, J.; Arendt-Nielsen, L. The Lancet Weight Determines Wheal Diameter in Response to Skin Prick Testing with Histamine. PLoS ONE 2016, 11, e0156211. [Google Scholar] [CrossRef]
- Marrugo, A.G.; Romero, L.A.; Pineda, J.; Vargas, R.; Altamar-Mercado, H.; Marrugo, J.; Meneses, J. Toward an Automatic 3D Measurement of Skin Wheals from Skin Prick Tests. In Proceedings of the Dimensional Optical Metrology and Inspection for Practical Applications VIII, Baltimore, MD, USA, 16–17 April 2019; International Society for Optics and Photonics. Volume 10991, p. 1099104. [Google Scholar]
- Pineda, J.; Vargas, R.; Romero, L.A.; Marrugo, J.; Meneses, J.; Marrugo, A.G. Robust Automated Reading of the Skin Prick Test via 3D Imaging and Parametric Surface Fitting. PLoS ONE 2019, 14, e0223623. [Google Scholar] [CrossRef]
- Rok, T.; Rokita, E.; Tatoń, G.; Guzik, T.; Śliwa, T. Thermographic Assessment of Skin Prick Tests in Comparison with the Routine Evaluation Methods. Postepy Dermatol. Alergol. 2016, 33, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Svelto, C.; Matteucci, M.; Pniov, A.; Pedotti, L. Skin Prick Test Digital Imaging System with Manual, Semiautomatic, and Automatic Wheal Edge Detection and Area Measurement. Multimed. Tools Appl. 2018, 77, 9779–9797. [Google Scholar] [CrossRef]
- Svelto, C.; Matteucci, M.; Resmini, R.; Pniov, A.; Pedotti, L.; Giordano, F. Semi-and-Automatic Wheal Measurement System for Prick Test Digital Imaging and Analysis. In Proceedings of the 2016 IEEE International Conference on Imaging Systems and Techniques (IST), Chania, Greece, 4–6 October 2016; pp. 482–486. [Google Scholar]
- Morales-Palacios, M.D.L.P.; Núñez-Córdoba, J.M.; Tejero, E.; Matellanes, Ó.; Quan, P.L.; Carvallo, Á.; Sánchez-Fernández, S.; Urtasun, M.; Larrea, C.; Íñiguez, M.T.; et al. Reliability of a Novel Electro-medical Device for Wheal Size Measurement in Allergy Skin Testing: An Exploratory Clinical Trial. Allergy 2023, 78, 299–301. [Google Scholar] [CrossRef]
- Uwitonze, J.P. Cost-Consequence Analysis of Computer Vision-Based Skin Prick Tests: Implications for Cost Containment in Switzerland. BMC Health Serv. Res. 2024, 24, 988. [Google Scholar] [CrossRef]
- Manca, D.P. Do Electronic Medical Records Improve Quality of Care? Yes. Can. Fam. Physician 2015, 61, 846–847, 850–851. [Google Scholar]
- Becker, A.S.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep Learning in Mammography: Diagnostic Accuracy of a Multipurpose Image Analysis Software in the Detection of Breast Cancer. Investig. Radiol. 2017, 52, 434–440. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; the CAMELYON16 Consortium; Hermsen, M.; Manson, Q.F.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; IEEE: Boston, MA, USA, 2015; pp. 3431–3440. [Google Scholar]
- Pena, J.C.; Pacheco, J.A.; Marrugo, A.G. Skin Prick Test Wheal Detection in 3D Images via Convolutional Neural Networks. In Proceedings of the 2021 IEEE 2nd International Congress of Biomedical Engineering and Bioengineering (CI-IB&BI), Virtual, 13–15 October 2021; IEEE: Bogota, Colombia, 2021. [Google Scholar]
- Lee, Y.H.; Shim, J.-S.; Kim, Y.J.; Jeon, J.S.; Kang, S.-Y.; Lee, S.P.; Lee, S.M.; Kim, K.G. Allergy Wheal and Erythema Segmentation Using Attention U-Net. J. Digit. Imaging. Inform. Med. 2024. [Google Scholar] [CrossRef]
- ImageMagick Studio LLC. ImageMagick, version 7.1.1-38. Available online: https://imagemagick.org (accessed on 1 October 2024).
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Taylor, L.; Nitschke, G. Improving Deep Learning Using Generic Data Augmentation. arXiv 2017, arXiv:1708.06020. [Google Scholar] [CrossRef]
- Ljanyst. image-segmentation-fcn. GitHub. 2017. Available online: https://github.com/ljanyst/image-segmentation-fcn (accessed on 1 October 2024).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A System for Large-Scale Machine Learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- OpenCV. OpenCV 4.1 Documentation. Available online: https://docs.opencv.org/4.1.0/ (accessed on 1 October 2024).
- Martin Bland, J.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 327, 307–310. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009; ISBN 1-4414-1269-7. [Google Scholar]
- Almeida, A.L.M.; Perger, E.L.P.; Gomes, R.H.M.; Sousa, G.d.S.; Vasques, L.H.; Rodokas, J.E.P.; Olbrich Neto, J.; Simões, R.P. Objective Evaluation of Immediate Reading Skin Prick Test Applying Image Planimetric and Reaction Thermometry Analyses. J. Immunol. Methods 2020, 487, 112870. [Google Scholar] [CrossRef] [PubMed]
- Serota, M.; Portnoy, J.; Jacobs, Z. Are Pseudopods On Skin Prick Testing Reproducible? J. Allergy Clin. Immunol. 2012, 129, AB239. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey 2020. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar]
- Punn, N.S.; Agarwal, S. Modality Specific U-Net Variants for Biomedical Image Segmentation: A Survey. Artif. Intell. Rev. 2022, 55, 5845–5889. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, R.H.M.; Perger, E.L.P.; Vasques, L.H.; Silva, E.G.M.D.; Simões, R.P. Deep Learning Method Applied to Autonomous Image Diagnosis for Prick Test. Life 2024, 14, 1256. https://doi.org/10.3390/life14101256
Gomes RHM, Perger ELP, Vasques LH, Silva EGMD, Simões RP. Deep Learning Method Applied to Autonomous Image Diagnosis for Prick Test. Life. 2024; 14(10):1256. https://doi.org/10.3390/life14101256
Chicago/Turabian StyleGomes, Ramon Hernany Martins, Edson Luiz Pontes Perger, Lucas Hecker Vasques, Elaine Gagete Miranda Da Silva, and Rafael Plana Simões. 2024. "Deep Learning Method Applied to Autonomous Image Diagnosis for Prick Test" Life 14, no. 10: 1256. https://doi.org/10.3390/life14101256






