In Situ Endothelial SARS-CoV-2 Presence and PROS1 Plasma Levels Alteration in SARS-CoV-2-Associated Coagulopathies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples Collection
2.2. Histology and Immunohistochemistry
2.3. Real-Time PCR
2.4. PROS1 ELISA Assay
2.5. Statistical Analysis
3. Results
3.1. Characterization of the Study Population
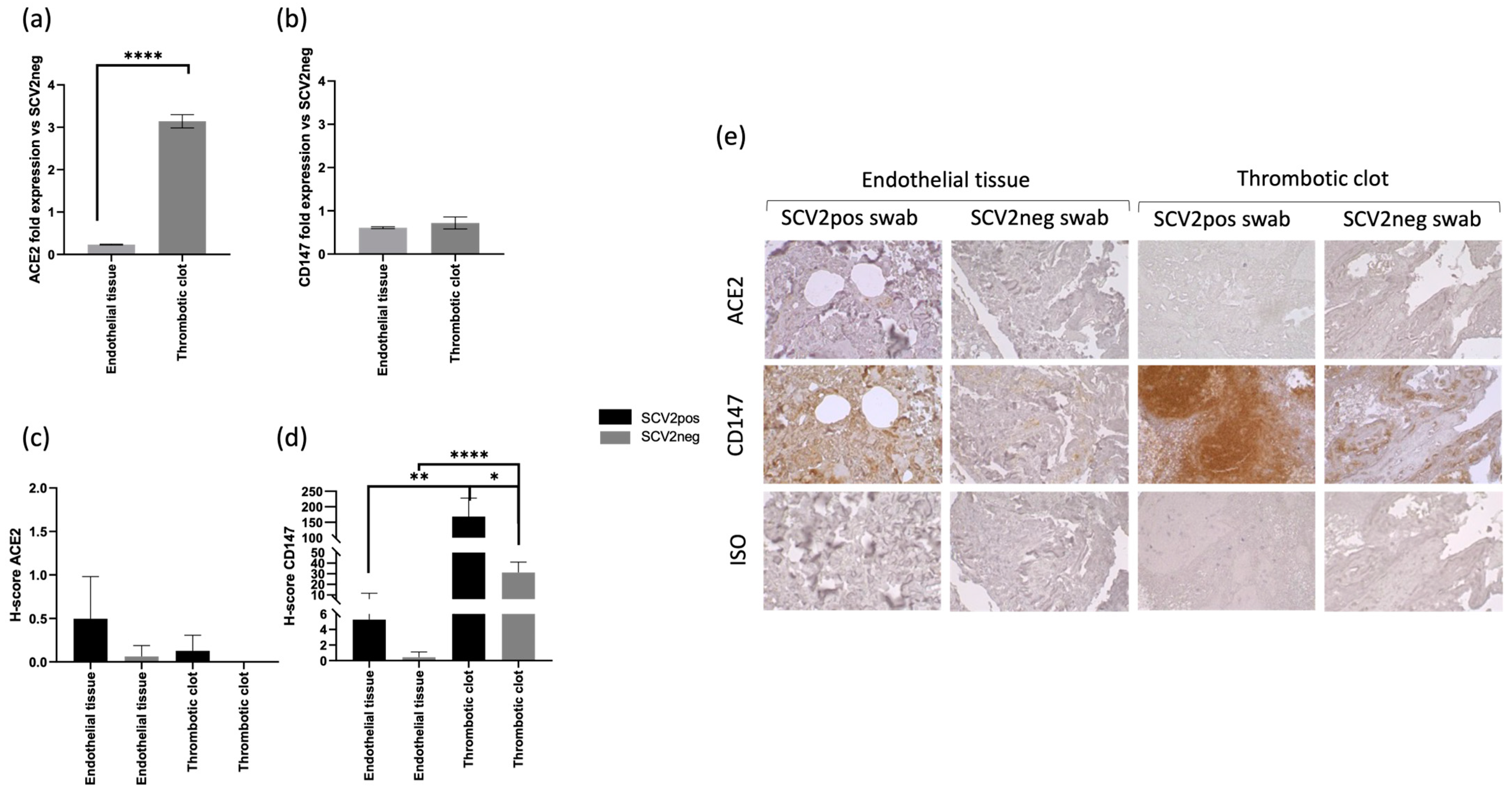
3.2. SARS-CoV-2 In Situ Presence Correlates with Previous Infection in Coagulopathic COVID-19 Subjects
3.3. CD147 Expression Correlates with SARS-CoV-2 In Situ Infection in Coagulopathic COVID-19 Subjects
3.4. SARS-CoV-2 In Situ Infection Correlates with Lower Plasma Levels of PROS1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Pohlmann, S. Novel SARS-CoV-2 receptors: ASGR1 and KREMEN1. Cell Res. 2022, 32, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Zhang, J.; Ma, X.; Tan, J.; Chen, L.; Liu, S.; Xin, Y.; Zhuang, L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 2020, 131, 110678. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Park, M.B.; Park, E.Y.; Lee, T.S.; Lee, J. Effect of the Period From COVID-19 Symptom Onset to Confirmation on Disease Duration: Quantitative Analysis of Publicly Available Patient Data. J. Med. Internet Res. 2021, 23, e29576. [Google Scholar] [CrossRef] [PubMed]
- Hartard, C.; Chaqroun, A.; Settembre, N.; Gauchotte, G.; Lefevre, B.; Marchand, E.; Mazeaud, C.; Nguyen, D.T.; Martrille, L.; Koscinski, I.; et al. Multiorgan and Vascular Tropism of SARS-CoV-2. Viruses 2022, 14, 515. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Lechner-Scott, J.; Levy, M.; Hawkes, C.; Yeh, A.; Giovannoni, G. Long COVID or post COVID-19 syndrome. Mult. Scler. Relat. Disord. 2021, 55, 103268. [Google Scholar] [CrossRef]
- López-León, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 Long-Term Effects of COVID-19: A Systematic Review and Meta-Analysis. SSRN Electron. J. 2021, 11, 16144. [Google Scholar] [CrossRef]
- Venkatesan, P. NICE guideline on long COVID. Lancet Respir. Med. 2021, 9, 129. [Google Scholar] [CrossRef]
- Bortolotti, D.; Simioni, C.; Neri, L.M.; Rizzo, R.; Semprini, C.M.; Occhionorelli, S.; Laface, I.; Sanz, J.M.; Schiuma, G.; Rizzo, S.; et al. Relevance of VEGF and CD147 in different SARS-CoV-2 positive digestive tracts characterized by thrombotic damage. FASEB J. 2021, 35, e21969. [Google Scholar] [CrossRef]
- Pretorius, E.; Vlok, M.; Venter, C.; Bezuidenhout, J.A.; Laubscher, G.J.; Steenkamp, J.; Kell, D.B. Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc. Diabetol. 2021, 20, 172. [Google Scholar] [CrossRef]
- Iba, T.; Levy, J.H.; Levi, M.; Connors, J.M.; Thachil, J. Coagulopathy of Coronavirus Disease 2019. Crit. Care Med. 2020, 48, 1358–1364. [Google Scholar] [CrossRef]
- Jose, R.J.; Manuel, A. COVID-19 cytokine storm: The interplay between inflammation and coagulation. Lancet Respir. Med. 2020, 8, e46–e47. [Google Scholar] [CrossRef]
- Ganier, L.; Morelli, X.; Borg, J.P. CD147 (BSG) but not ACE2 expression is detectable in vascular endothelial cells within single cell RNA sequencing datasets derived from multiple tissues in healthy individuals. Med. Sci. 2020, 36, 42–46. [Google Scholar] [CrossRef]
- Schimmel, L.; Chew, K.Y.; Stocks, C.J.; Yordanov, T.E.; Essebier, P.; Kulasinghe, A.; Monkman, J.; Dos Santos Miggiolaro, A.F.R.; Cooper, C.; de Noronha, L.; et al. Endothelial cells are not productively infected by SARS-CoV-2. Clin. Transl. Immunol. 2021, 10, e1350. [Google Scholar] [CrossRef] [PubMed]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Spadaro, S.; Strazzabosco, G.; Campo, G.; Carosella, E.D.; Papi, A.; et al. Increased sHLA-G Is Associated with Improved COVID-19 Outcome and Reduced Neutrophil Adhesion. Viruses 2021, 13, 1855. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.; Favaloro, E.J.; Lippi, G.; Van Cott, E.M. Hematology Laboratory Abnormalities in Patients with Coronavirus Disease 2019 (COVID-19). Semin. Thromb. Hemost. 2020, 46, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Y.; Wang, X.; Yang, L.; Li, H.; Wang, Y.; Liu, M.; Zhao, X.; Xie, Y.; Yang, Y.; et al. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J. Hematol. Oncol. 2020, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Pennings, G.J.; Yong, A.S.; Kritharides, L. Expression of EMMPRIN (CD147) on circulating platelets in vivo. J. Thromb. Haemost. 2010, 8, 472–481. [Google Scholar] [CrossRef]
- Tutusaus, A.; Mari, M.; Ortiz-Perez, J.T.; Nicolaes, G.A.F.; Morales, A.; Garcia de Frutos, P. Role of Vitamin K-Dependent Factors Protein S and GAS6 and TAM Receptors in SARS-CoV-2 Infection and COVID-19-Associated Immunothrombosis. Cells 2020, 9, 2186. [Google Scholar] [CrossRef]
- Castoldi, E.; Hackeng, T.M. Regulation of coagulation by protein S. Curr. Opin. Hematol. 2008, 15, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, J.A. Identification of the antithrombotic protein S as a potential target of the SARS-CoV-2 papain-like protease. Thromb. Res. 2020, 196, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Wypasek, E.; Karpinski, M.; Alhenc-Gelas, M.; Undas, A. Venous thromboembolism associated with protein S deficiency due to Arg451* mutation in PROS1 gene: A case report and a literature review. J. Genet. 2017, 96, 1047–1051. [Google Scholar] [CrossRef]
- Baez-Santos, Y.M.; St John, S.E.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Pilli, V.S.; Plautz, W.; Majumder, R. The Journey of Protein S from an Anticoagulant to a Signaling Molecule. JSM Biochem. Mol. Biol. 2016, 3, 1014. [Google Scholar]
- Ziebuhr, J. The coronavirus replicase. Curr. Top Microbiol. Immunol. 2005, 287, 57–94. [Google Scholar] [CrossRef]
- Zaid, Y.; Puhm, F.; Allaeys, I.; Naya, A.; Oudghiri, M.; Khalki, L.; Limami, Y.; Zaid, N.; Sadki, K.; Ben El Haj, R.; et al. Platelets Can Associate with SARS-CoV-2 RNA and Are Hyperactivated in COVID-19. Circ. Res. 2020, 127, 1404–1418. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Strazzabosco, G.; Fernandez, M.; Caccuri, F.; Caruso, A.; Rizzo, R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms 2021, 9, 1820. [Google Scholar] [CrossRef]
- Baroni, M.; Pavani, G.; Marescotti, D.; Kaabache, T.; Borgel, D.; Gandrille, S.; Marchetti, G.; Legnani, C.; D’Angelo, A.; Pinotti, M.; et al. Membrane binding and anticoagulant properties of protein S natural variants. Thromb. Res. 2010, 125, e33–e39. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Zamboni, P.; Bortolotti, D.; Occhionorelli, S.; Traina, L.; Neri, L.M.; Rizzo, R.; Gafa, R.; Passaro, A. Bowel ischemia as onset of COVID-19 in otherwise asymptomatic patients with persistently negative swab. J. Intern. Med. 2022, 291, 224–231. [Google Scholar] [CrossRef]
- Kumar, A.; Narayan, R.K.; Kumari, C.; Faiq, M.A.; Kulandhasamy, M.; Kant, K.; Pareek, V. SARS-CoV-2 cell entry receptor ACE2 mediated endothelial dysfunction leads to vascular thrombosis in COVID-19 patients. Med. Hypotheses 2020, 145, 110320. [Google Scholar] [CrossRef]
- Beyerstedt, S.; Casaro, E.B.; Rangel, E.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Pennings, G.J.; Kritharides, L. CD147 in cardiovascular disease and thrombosis. Semin. Thromb. Hemost. 2014, 40, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells 2021, 10, 1434. [Google Scholar] [CrossRef]
- Maugeri, N.; De Lorenzo, R.; Clementi, N.; Antonia Diotti, R.; Criscuolo, E.; Godino, C.; Tresoldi, C.; Angels For COVID-Bio, B.S.G.B.; Bonini, C.; Clementi, M.; et al. Unconventional CD147-dependent platelet activation elicited by SARS-CoV-2 in COVID-19. J. Thromb. Haemost. 2022, 20, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Ghasemzadeh, M.; Ahmadi, J.; Hosseini, E. Platelet-leukocyte crosstalk in COVID-19: How might the reciprocal links between thrombotic events and inflammatory state affect treatment strategies and disease prognosis? Thromb. Res. 2022, 213, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.D.; Aceves, N.M.; Liu, J.L.; Compton, J.R.; Leary, D.H.; Freitas, B.T.; Pegan, S.D.; Doctor, K.Z.; Wu, F.Y.; Hu, X.; et al. The SARS-CoV-2 SSHHPS Recognized by the Papain-like Protease. ACS Infect. Dis. 2021, 7, 1483–1502. [Google Scholar] [CrossRef] [PubMed]



| SARS-CoV-2 Positive Swab (N = 7) | SARS-CoV-2 Negative Swab (N = 11) | p Values | |
|---|---|---|---|
| Gender; N; % | M (5; 71%) F (2; 29%) | M (9; 82%) F (2; 18%) | 0.51 |
| Age; mean ± SD | 85.0 ± 3.3 | 70.8 ± 6.7 | <0.0001 |
| Thromboendarterectomy | 57% (4) | 55% (6) | 0.65 |
| Amputation | 29% (2) | 0% (0) | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baroni, M.; Beltrami, S.; Schiuma, G.; Ferraresi, P.; Rizzo, S.; Passaro, A.; Molina, J.M.S.; Rizzo, R.; Di Luca, D.; Bortolotti, D. In Situ Endothelial SARS-CoV-2 Presence and PROS1 Plasma Levels Alteration in SARS-CoV-2-Associated Coagulopathies. Life 2024, 14, 237. https://doi.org/10.3390/life14020237
Baroni M, Beltrami S, Schiuma G, Ferraresi P, Rizzo S, Passaro A, Molina JMS, Rizzo R, Di Luca D, Bortolotti D. In Situ Endothelial SARS-CoV-2 Presence and PROS1 Plasma Levels Alteration in SARS-CoV-2-Associated Coagulopathies. Life. 2024; 14(2):237. https://doi.org/10.3390/life14020237
Chicago/Turabian StyleBaroni, Marcello, Silvia Beltrami, Giovanna Schiuma, Paolo Ferraresi, Sabrina Rizzo, Angelina Passaro, Juana Maria Sanz Molina, Roberta Rizzo, Dario Di Luca, and Daria Bortolotti. 2024. "In Situ Endothelial SARS-CoV-2 Presence and PROS1 Plasma Levels Alteration in SARS-CoV-2-Associated Coagulopathies" Life 14, no. 2: 237. https://doi.org/10.3390/life14020237
APA StyleBaroni, M., Beltrami, S., Schiuma, G., Ferraresi, P., Rizzo, S., Passaro, A., Molina, J. M. S., Rizzo, R., Di Luca, D., & Bortolotti, D. (2024). In Situ Endothelial SARS-CoV-2 Presence and PROS1 Plasma Levels Alteration in SARS-CoV-2-Associated Coagulopathies. Life, 14(2), 237. https://doi.org/10.3390/life14020237










