Fibrin and Fibrinolytic Enzyme Cascade in Thrombosis: Unravelling the Role
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
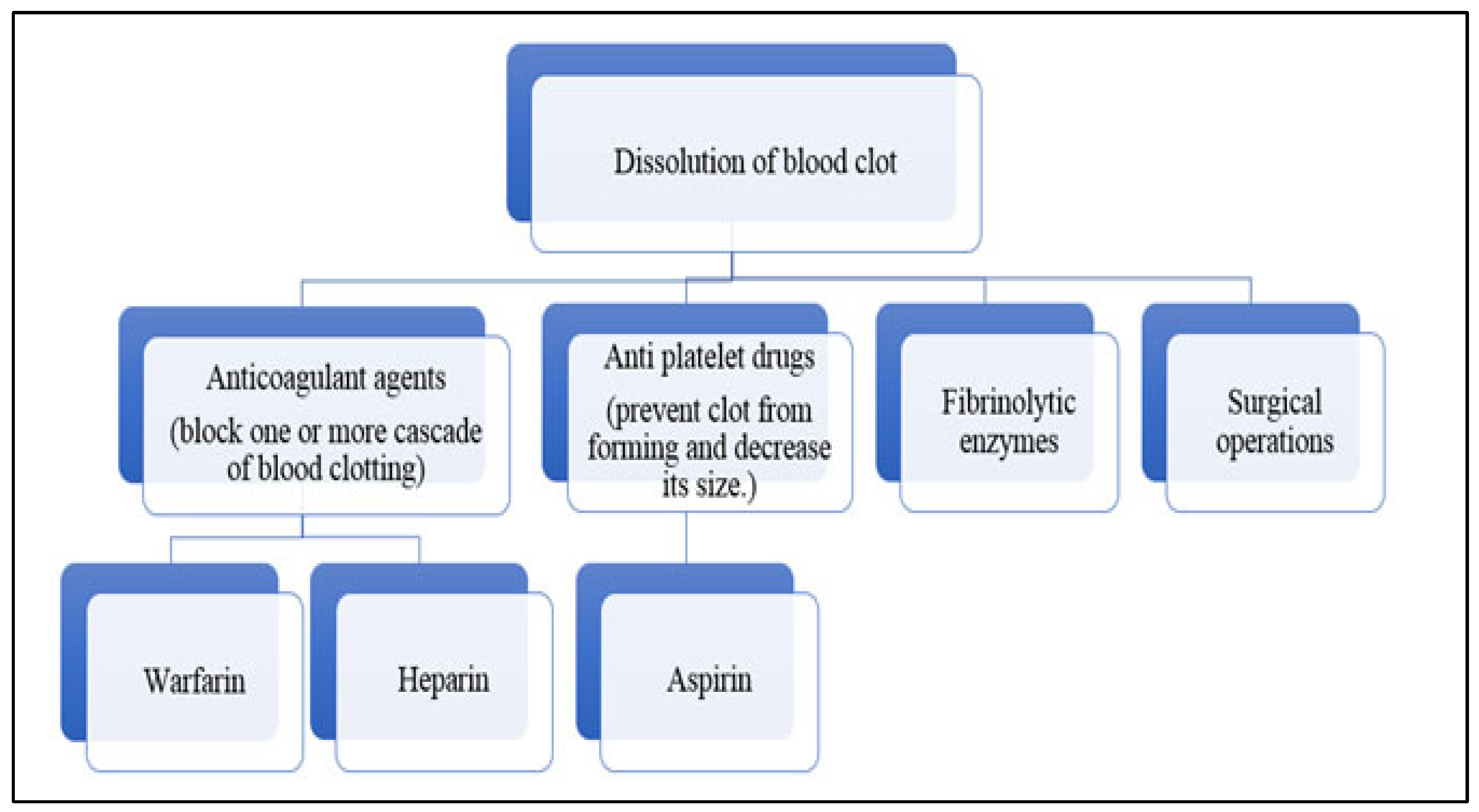
3.1. Cardiovascular Diseases and Thrombosis
3.2. Molecular Mechanism of Clot Formation
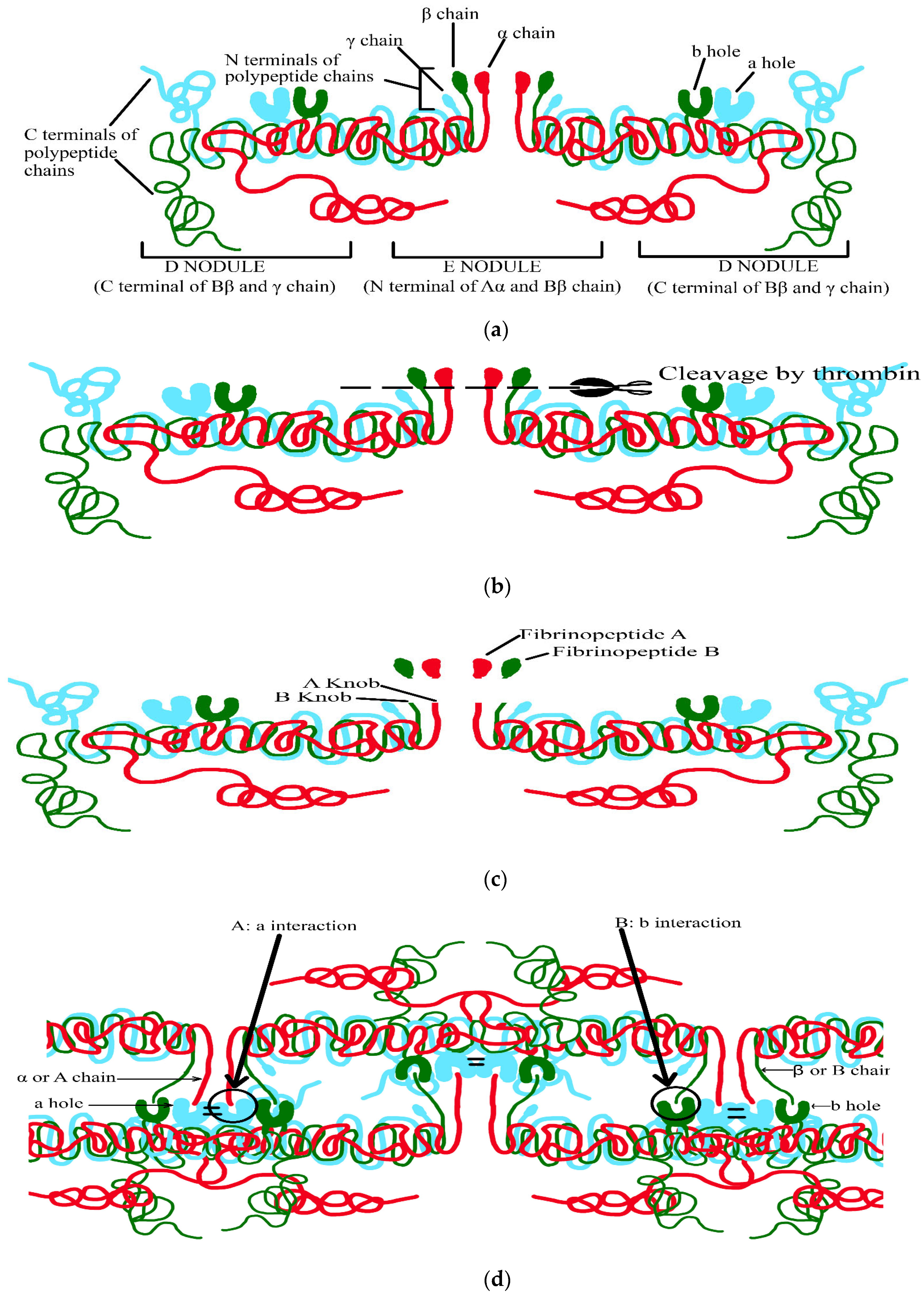
- (1)
- The N terminal of the E nodule,
- (2)
- The C terminal of the Υ- and βB-chains from the D nodule facing outwards,
- (3)
- (1)
- Thrombin attaches to central E nodule cleaving N terminal peptides of Aα- and Bβ-chains (Figure 2b).
- (2)
- Aα-chains are firstly cleaved by thrombin at a faster rate releasing fibrinopeptide A containing N terminal (16 residues), exposing the binding site containing Gly-Pro-Arg in E region (A knob) (Figure 2c).
- (3)
- A knob has a complementary binding site of Υ-chain D region (a hole) creating (A: a) interaction mediating the formation of protofibrils, which are metastable peptide assemblies observed during the growth of amyloid fibrils by a number of peptides (Figure 2d)
- (4)
- Subsequently, removal of fibrinopeptide B containing N terminal (14 residues) causes a release, exposing the binding site containing Gly-His-Arg in E region (B knob) (Figure 2c).
- (5)
- B knob also has a complementary binding site of β chain D region (b hole) creating (B: b) interaction, thus, mediating the lateral aggregation of fibrinogen (Figure 2d).
3.3. Fibrin Architecture
- (1)
- Fibrin is formed in vivo at the site of blood vessel lesion where platelets are stimulated and bind to fibrin forming powerful adhesive forces. These fibres under tension regulate clot structure, constrain fibrin fibres, and increase their density in platelet-rich areas [36].
- (2)
- Factor XIIIa (transglutaminase) proposes Υ-glutamyllysine crosslinking between αC- and ΥC-domains of next fibrin monomers and tightens up the lateral (flanking) attachment of protofibrils. This covalent crosslinking results in a decrease in the fibrin diameter without any modification in the number of protofibrils in fibres. Thus, decreasing the vacant fluid space volume within the fibres causes a two-fold reduction in pore size [37,38].
- (3)
- DNA and histones that are released by activated neutrophils form neutrophil extracellular traps. These have a major effect on clot lysis as they hold lysing fibrin (large fibrin degradative products) together resulting in a delay in the fibrinolytic process [39].
- (4)
- The contractile force (induced by fibrin) of neighbouring fibres activates platelets which acts on red blood cells (RBCs) causing a change in their configuration from biconcave to polyhedral. This change induces gap-free compression of RBCs in unoccupied spaces between fibrin fibres forming a high lytic resistance structure and strong diffusion barrier [40]. These activities of RBCs increase blood viscosity, and express phosphatidylserine on their surface, which promotes fibrin deposition during venous thrombosis and reduces clot dissolution by suppressing plasmin [31].
3.4. Fibrinolysis
- (1)
- Tissue-type plasminogen activator (t-PA) which is enhanced in the presence of fibrin.
- (2)
- Urokinase-type plasminogen activator (u-PA), which binds to specific u-PA receptors, enhancing the activation of cell-bound plasminogen (Figure 3).
Components of the Fibrinolytic System
- (1)
- Plasminogen
- Heavy chains have an N-terminal part of plasminogen including five kringles.
- Light chains having the C-terminal part of plasminogen containing serine peptidase (catalytic triad: His-603, Asp-646, Ser-741) [38].
- (2)
- Tissue-type plasminogen activator (t-PA).
- (3)
- Urokinase
- (4)
- Plasmin
- Plasmin deactivates and cleaves various clotting factors FV, FVIII, FIX, and FX in vitro which plays a major role in clot formation [57].
- The two catalytic A-subunits of active clotting factor XIII are degraded endogenously by plasmin during lysis of the blood clot [58].
- Plasmin is an important matrix metalloprotease activator, enhancing the lysis effect of plasmin on surrounding tissues [59].
- (5)
- Plasminogen activator inhibitor
- (6)
- α2-Antiplasmin
- Inhibiting the adsorption of plasminogen to fibrin: the C-terminal end of α2-antiplasmin binds with a robust affinity towards the lysine binding site, where fibrin is bound non-covalently (competitive inhibition).
- Formation of a balanced inactive complex by plasmin: after the binding of α2-antiplasmin with the lysine binding site, it is quickly cleaved via plasmin at the active site releasing the peptides and forming a covalent plasmin—α2-antiplasmin complex.
- Cross-linkage via factor XIIIa: the portion of circulating α2-antiplasmin is tightly bound to fibrin via factor XIIIa, resulting in the amplified resistance of fibrin to fibrinolysis.
3.5. Why Measure Fibrinolysis?
3.6. Fibrinolytic Activity Assay
- (1)
- Fibrin plate assay
- Plasminogen-free fibrin plate (heated): This assay allows the direct activity of plasmin-like enzymes, formed from fibrinogen solution (5 mg human fibrinogen in 7 mL of 0.1 M Barbital buffer of 7.8 pH), 10 U thrombin solution and 7 mL of 10 g agarose/Liter) to be assessed in Petri plates. Then, for inactivating fibrinolytic enzymes, the plates were heated at 80 °C for 30 min. These plates were modified by means of bovine fibrinogen, calcium chloride, thrombin, and sodium chloride. The enzyme (10–30 µL) was dropped judiciously on a fibrin plate and incubated at 37 °C temperature for 3–18 h, and clear zones were obtained. A standard curve was plotted by using standard fibrinolytic enzyme (urokinase) to examine the fibrinolytic activity of an enzyme.
- Plasminogen-rich fibrin plate (non-heated): This consists of 5 U plasminogen in addition to the above fibrinolytic solution and is not heated. It is suitable for plasminogen activators [66].
- (2)
- Fibrin microplate assay
- (3)
- Rapid fibrin plate assay
- (4)
- Euglobulin clot lysis time (ECLT)
- (5)
- Global fibrinolytic capacity (GFC)
- (6)
- Viscoelastic method
- Rotational thromboelastometry (ROTEM) computes different viscoelastic parameters like clotting time, clot growth kinetics, the pace of coagulation initiation, clot strength, and dissolution [66]. The five principal assays used with the ROTEM instrument are INTEM, HEPTEM, EXTEM, FIBTEM, and APTEM assays. The INTEM test initiates clotting via the intrinsic pathway using ellagic acid, while the HEPTEM assay uses heparinase in addition to ellagic acid. EXTEM uses tissue factor to initiate the extrinsic clotting cascade whereas FIBTEM uses cytochalasin D to inhibit platelet activity and provide clot tracing that indicates the presence of fibrinogen. This test is used extensively in cardiac and liver studies to monitor fibrinogen levels. APTEM is a modified EXTEM assay that incorporates aprotinin to stabilize the clot against hyperfibrinolysis. An electrical signal from an automatic electrical transducer leads to a graphical display supervised by a computer [71,72].
- Thromboelastography (TEG) is a non-invasive test that quickly determines coagulation rate (hypo/hyper) or solidification to fibrinolysis (involving prothrombin/thrombin/fibrin), the viscoelastic properties of blood samples during clotting under low shear stress. It uses reagents different from ROTEM and involves five different parameters: reaction time, kinetics, alpha angle, maximum amplitude, and lysis at 30 min (A30/LY30) [5,72].
- Sonoclot: This assesses the change in resistivity via a small disposable plastic probe spinning vertically on a coagulating blood sample in the cuvette. Fibrin components formed on the tip/ around the probe and on the internal wall of the cuvette increase the weight of the probe leading to an upsurge in the resistivity. This increase in resistivity is sensed via electronic circuits and transformed into an output signal. The output signal describes the viscoelastic properties of the blood coagulation initiated from fibrin development, aggregation of fibrin monomers, platelet interaction, clot retraction, and lysis [73].
3.7. Microorganisms: Important Source of Fibrinolytic Enzymes
- Endopeptidase: Serine protease: trypsin, thrombin, chymotrypsin, subtilisin, etc.Cysteine protease: rhinovirus 3C, papain, etc.Metalloprotease: collagenase, thermolysin, etc.Aspartic protease: pepsin and cathepsin [83].
- Exopeptidase: Serine protease: carboxypeptidase Y.Cystine protease: cathepsin and DAPase.Metalloprotease: carboxypeptidase A, carboxypeptidase B [83].Serine and metalloprotease have catalytic properties of fibrinolytic enzymes.
3.8. Nattokinase (NK)
3.9. Streptokinase (SK)
3.10. Staphylokinase (SAK)
3.11. Serrapeptase (SRP)
3.12. Longolytin
3.13. Clinical Significance of Fibrinolytic Enzymes
3.14. Other Potential Applications of Fibrinolytic Enzymes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- MFMER: Rochester, MN, USA. Available online: https://www.mayoclinic.org/diseases-conditions/heart-disease/symptoms-causes/syc-20353118 (accessed on 15 September 2023).
- Sharma, C.; Osmolovskiy, A.; Singh, R. Microbial Fibrinolytic Enzymes as Anti-Thrombotics: Production, Characterisation and Prodigious Biopharmaceutical Applications. Pharmaceutics 2021, 13, 1880. [Google Scholar] [CrossRef] [PubMed]
- Geddings, J.E.; Mackamn, N. Recently identified factors that regulate hemostatsis and thrombosis. Thromb. Haemost. 2014, 111, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.merriam-webster.com/dictionary/thromboembolism (accessed on 15 September 2023).
- Longstaff, C. Measuring fibrinolysis: From research to routine diagnostic assays. J. Thromb. Haemost. 2018, 16, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Kotb, E. The Biotechnological Potential of Fibrinolytic Enzymes in the Dissolution of Endogenous Blood Thrombi. Biotechnol. Prog. 2014, 30, 656–672. [Google Scholar] [CrossRef]
- Marsh, N.A.; Gaffney, P.J. A rapid fibrin plate: A method for plasminogen activator assay. Thromb. Haemost. 1977, 38, 545–551. [Google Scholar] [CrossRef]
- Hmidet, N.; Nawani, N.; Ghorbel, S. Recent Development in Production and Biotechnological Application of Microbial Enzymes. Biomed. Res. Int. 2014, 2015, 2. [Google Scholar] [CrossRef]
- Liang, X.; Jia, S.; Sun, Y.; Chen, M.; Chen, X.; Zhong, J.; Huan, L. Secretory Expression of Nattokinase from Bacillus subtilis YF38 in Escherichia coli. Mol. Biotechnol. 2007, 37, 187–194. [Google Scholar] [CrossRef]
- Dimmeler, S. Cardiovascular diseases review series. EMBO Mol. Med. 2011, 3, 697. [Google Scholar] [CrossRef]
- Lijfering, W.; Flinterman, L.; Vandenbroucke, J.; Rosendaal, F.; Cannegieter, S. Relationship between Venous and Arterial Thrombosis: A Review of the Literature from a Causal Perspective. Semin. Thromb. Hemost. 2011, 37, 885–896. [Google Scholar] [CrossRef]
- Lijnen, H.R.; Collen, D. Impaired fibrinolysis and the risk for coronary heart disease. Circulation 1996, 94, 2052–2054. [Google Scholar] [CrossRef]
- Unar, A.; Bertolino, L.; Patauner, F.; Gallo, R.; Durante-Mangoni, E. Pathophysiology of Disseminated Intravascular Coagulation in Sepsis: A Clinically Focused Overview. Cells 2023, 12, 2120. [Google Scholar] [CrossRef]
- Unar, A.; Bertolino, L.; Patauner, F.; Gallo, R.; Durante-Mangoni, E. Decoding Sepsis-Induced Disseminated Intravascular Coagulation: A Comprehensive Review of Existing and Emerging Therapies. J. Clin. Med. 2023, 12, 6128. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.; Meade, D.M. Thrombosis and cardiovascular disease. Med. Clin. N. Am. 1992, 82, 511–522. [Google Scholar]
- Nabel, E.G. Cardiovascular disease. N. Engl. J. Med. 2003, 349, 60–72. [Google Scholar] [CrossRef]
- Bagot, C.N.; Arya, R. Virchow and his triad: A question of attribution. Br. J. Haematol. 2008, 143, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Del Principe, M.I.; Del Principe, D.; Venditti, A. Thrombosis in adult patients with acute leukemia. Curr. Opin. Oncol. 2017, 29, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Guo, H.; Zhang, Y.; Qiao, R. Procoagulant platelets: Generation, characteristics, and therapeutic target. J. Clin. Lab. Anal. 2021, 35, e23750. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Salem, G.E.M.; Sharma, N.; Gautam, P.; Singh, R. Thrombolytic Potential of Novel Thiol-Dependent Fibrinolytic Protease from Bacillus cereus RSA1. Biomolecules 2020, 10, 3. [Google Scholar] [CrossRef]
- Watson, L.; Broderick, C.; Armon, M.P. Thrombolysis for acute deep vein thrombosis. Cochrane Database Syst. Rev. 2016, 11, CD002783. [Google Scholar] [CrossRef]
- Nagareddy, P.; Smyth, S.S. Inflammation and thrombosis in cardiovascular disease. Curr. Opin. Hematol. 2013, 20, 457–463. [Google Scholar] [CrossRef]
- Chapin, J.C.; Hajjar, K.A. Fibrinolysis and the control of blood coagulation. Blood Rev. 2015, 29, 17–24. [Google Scholar] [CrossRef]
- La Corte, A.L.C.; Philippou, H.; Ariens, R.A.S. Role of fibrin structure in thrombosis and vascular disease. Adv. Protein Chem. Struct. Biol. 2011, 83, 75–127. [Google Scholar]
- Lord, S.T. Fibrinogen and fibrin: Scaffold proteins in haemostasis. Curr. Opin. Hematol. 2007, 14, 236–241. [Google Scholar] [CrossRef]
- Schuligaa, M.; Graingeb, C.; Westall, G.; Knigh, D. The fibrogenic actions of the coagulant and plasminogen activation systems in pulmonary fibrosis. Int. J. Biochem. Cell Biol. 2018, 97, 108–117. [Google Scholar] [CrossRef]
- Wolberg, A.S. Thrombin generation and fibrin clot structure. Blood Rev. 2007, 21, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.V.; Standeven, K.F.; Arie, R.A.S. Fibrinogen gamma-chain splice variant alters fibrin formation and structure. Blood 2003, 102, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Bagoly, Z.; Koncz, Z.; Hársfalvi, J.; Muszbek, L. Factor XIII, clot structure, thrombosis. Thromb. Res. 2011, 129, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Ariens, R.A.S. Fibrin (ogen) and Thrombotic disease. J. Thromb. Haemost. 2013, 11, 294–305. [Google Scholar] [CrossRef]
- Aleman, M.M.; Walton, B.L.; Byrnes, J.R.; Wolberg, A.S. Fibrinogen and red blood cells in venous thrombosis. Thromb. Res. 2014, 133, S38–S40. [Google Scholar] [CrossRef] [PubMed]
- Chernysh, I.N.; Nagaswami, C.; Weisel, J.W. Visualization and identification of the structures formed during early stages of fibrin polymerization. Blood 2016, 117, 4609–4614. [Google Scholar] [CrossRef]
- Blombäck, B.; Carlsson, K.; Fatah, K.; Hessel, B.; Procyk, R. Fibrin in human plasma: Gel architectures governed by rate and nature of fibrinogen activation. Thromb. Res. 1994, 75, 521–538. [Google Scholar] [CrossRef]
- Bridge, K.I.; Philippou, H.; Ariëns, R.A.S. Clot properties and cardiovascular disease. Thromb. Haemost. 2014, 112, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Collet, J.P.; Lesty, C.; Montalescot, G.; Weisel, J.W. Dynamic Changes of Fibrin Architecture during Fibrin Formation and Intrinsic Fibrinolysis of Fibrin-rich Clots. J. Biol. Chem. 2013, 278, 21331–21335. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.A.; Chaudhuri, O.; Crow, A.; Webster, K.D.; Li, T.-D.; Kita, A.; Huang, J.; Fletcher, D.A. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nat. Mater. 2011, 10, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.A.; Mockros, L.F.; Weisel, J.W.; Lorand, L. Structural Origins of Fibrin Clot Rheology. Biophys. J. 1999, 77, 2813–2826. [Google Scholar] [CrossRef]
- Hethershaw, E.L.; La Corte, A.C.; Duval, C.; Ali, M.; Grant, P.J.; Ariëns, R.A.; Philippou, H. The effect of blood coagulation factor XIII on fibrin clot structure and fibrinolysis. J. Thromb. Haemost. 2014, 12, 197–205. [Google Scholar] [CrossRef]
- Longstaff, C.; Varjú, I.; Sótonyi, P.; Szabó, L.; Krumrey, M.; Hoell, A.; Bóta, A.; Varga, Z.; Komorowicz, E.; Kolev, K. Mechanical Stability and Fibrinolytic Resistance of Clots Containing Fibrin, DNA, and Histones. J. Biol. Chem. 2013, 288, 6946–6956. [Google Scholar] [CrossRef]
- Longstaff, C.; Kolev, K. Basic mechanisms and regulation of fibrinolysis. J. Thromb. Haemost. 2015, 13, S98–S105. [Google Scholar] [CrossRef]
- Sharma, C.; Nigam, A.; Singh, R. Computational-approach understanding the structure-function prophecy of Fibrinolytic Protease RFEA1 from Bacillus cereus RSA1. PeerJ 2021, 9, e11570. [Google Scholar] [CrossRef]
- Rijken, D.J.; Lijnen, H.R. New insights into the molecular mechanisms of the fibrinolytic system. J. Thromb. Haemost. 2009, 7, 4–13. [Google Scholar] [CrossRef]
- Gabriel, D.A.; Muga, K.; Boothroyd, E.M. The Effect of Fibrin Structure on Fibrinolysis. J. Biol. Chem. 1992, 267, 24259–24263. [Google Scholar] [CrossRef] [PubMed]
- Hoylaerts, M.; Rijken, D.C.; Lijnen, H.R.; Collen, D. Kinetics of the Activation of Plasminogen by Human Tissue Plasminogen Activator. Role of Fibrin. J. Biol. Chem. 1982, 257, 2912–2919. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.J. Role of tissue-type plasminogen activator and plasminogen activator inhibitor-1 in psychological stress and depression. Oncotarget 2017, 8, 113258–113268. [Google Scholar] [CrossRef] [PubMed]
- Rijken, D.C.; Hoylaerts, M.; Collen, D. Fibrinolytic Properties of One-chain and Two-chain Human Extrinsic (Tissue-type) Plasminogen Activator. J. Biol. Chem. 1982, 257, 2920–2925. [Google Scholar] [CrossRef] [PubMed]
- Longstaff, C.; Thelwell, C.; Williams, S.C.; Silva, M.M.; Szabó, L.; Kolev, K. The interaction between tissue plasminogen activator domains and fibrin structure studies in the regulation of fibrinolysis: Kinetic and microscopic structure. Blood 2010, 117, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.J.; Leonard, C.K.; Guzzetta, A.W.; Spellman, M.W. Tissue plasminogen activator has an O-linked fucose attached to threonine-61 in the epidermal growth factor domain. Biochem. J. 1991, 30, 2311–2314. [Google Scholar] [CrossRef]
- Cesarman-Maus, G.; Hajjar, K.A. Molecular mechanisms of fibrinolysis. Br. J. Haematol. 2005, 129, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Munk, G.A.W.; Groeneveld, E.; Rijken, D.C. Acceleration of the Thrombin Inactivation of Single Chain Urokinase-type Plasminogen Activator (Pro-urokinase) by Thrombomodulin. J. Clin. Investig. 1991, 88, 1680–1684. [Google Scholar] [CrossRef]
- Liu, J.N.; Gurewich, V. The kinetics of plasminogen activation by thrombin-cleaved pro-urokinase and promotion of its activity by fibrin fragment E-2 and by tissue plasminogen activator. Blood 1993, 81, 980–987. [Google Scholar] [CrossRef]
- Ploug, M.; Ellis, V. Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom a-neurotoxins. FEBS Lett. 1994, 349, 163–168. [Google Scholar] [CrossRef]
- Bohuslav, J.; Horejsí, V.; Hansmann, C.; Stöckl, J.; Weidle, U.H.; Majdic, O.; Bartke, I.; Knapp, W.; Stockinger, H. Urokinase Plasminogen Activator Receptor, 2-Integrins, and Src-kinases within a Single Receptor Complex of Human Monocytes. J. Exp. Med. 1995, 181, 1381–1390. [Google Scholar] [CrossRef]
- Gurewich, V.; Pannell, R.; Louie, S.; Kelley, P.; Suddith, R.L.; Greenlee, R. Effective and Fibrin-specific Clot Lysis by a Zymogen Precursor Form of Urokinase (Pro-urokinase) A Study In Vitro and in Two Animal Species. J. Clin. Investig. 1984, 73, 1731–1739. [Google Scholar] [CrossRef]
- Lijnen, H.R.; Zamarron, C.; Blaber, M.; Winkler, M.E.; Collen, D. Activation of Plasminogen by Pro-urokinase. J. Biol. Chem. 1985, 261, 1253–1258. [Google Scholar] [CrossRef]
- Schneider, M.; Brufatto, N.; Neill, E.; Nesheim, M. Activated Thrombin-activatable Fibrinolysis Inhibitor Reduces the Ability of High Molecular Weight Fibrin Degradation Products to Protect Plasmin from Antiplasmin. J. Biol. Chem. 2004, 279, 13340–13345. [Google Scholar] [CrossRef] [PubMed]
- Hoover-Plow, J. Does plasmin have anticoagulant activity? Vasc. Health Risk Manag. 2010, 6, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Hur, W.S.; Mazinani, N.; Lu, X.J.D.; Britton, H.M.; Byrnes, J.R.; Wolberg, A.S.; Kastrup, C.J. Coagulation factor XIIIa is inactivated by plasmin. Blood 2015, 126, 2329–2337. [Google Scholar] [CrossRef] [PubMed]
- Bokarewa, M.I.; Jin, T.; Tarkowski, A. Staphylococcus aureus: Staphylokinase. Int. J. Biochem. Cell Biol. 2005, 38, 504–509. [Google Scholar] [CrossRef]
- Nar, H.; Bauer, M.; Stassen, J.-M.; Lang, D.; Gils, A.; Declerck, P.J. Plasminogen Activator Inhibitor 1. Structure of the Native Serpin, Comparison to its Other Conformers and Implications for Serpin Inactivation. J. Mol. Biol. 2000, 297, 683–695. [Google Scholar] [CrossRef]
- Pant, A.; Kopec, A.K.; Baker, K.S.; Cline-Fedewa, H.; Lawrence, D.A.; Luyendyk, J.P. Plasminogen Activator Inhibitor-1 Reduces tPA-Dependent Fibrinolysis and Intrahepatic Hemorrhage in Experimental Acetaminophen Overdose. Am. J. Pathol. 2018, 188, 1204–1212. [Google Scholar] [CrossRef]
- Masson, C.; Angles-Cano, E. Kinetic analysis of the interaction between plasminogen activator inhibitor-1 and tissue-type plasminogen activator. Biochem. J. 1988, 256, 237–244. [Google Scholar] [CrossRef]
- Zakrzewski, M.; Zakrzewska, E.; Kiciński, P.; Przybylska-Kuć, S.; Dybała, A.; Myśliński, W.; Pastryk, J.; Tomaszewski, T.; Mosiewicz, J. Evaluation of Fibrinolytic Inhibitors: Alpha-2 Antiplasmin and Plasminogen Activator Inhibitor 1 in Patients with Obstructive Sleep Apnoea. PLoS ONE 2016, 11, e0166725. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.L.; Mathew, P. α2-Antiplasmin and its deficiency: Fibrinolysis out of balance. Haemophilia 2008, 16, 1250–1254. [Google Scholar] [CrossRef]
- Ilich, A.; Bokarev, I.; Key, N.S. Global assays of fibrinolysis. Int. J. Lab. Hematol. 2017, 39, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Kotb, E. Activity assessment of microbial fibrinolytic enzymes. Appl. Microbiol. Biotechnol. 2013, 97, 6647–6665. [Google Scholar] [CrossRef] [PubMed]
- Fossum, S.; Hoem, N.O. Urokinase and non-urokinase fibrinolytic activity in protease-inhibitor-deprived plasma, assayed by a fibrin micro-plate method. Immunopharmacology 1996, 32, 119–121. [Google Scholar] [CrossRef]
- Boudjeltia, K.Z.; Cauchie, P.; Remacle, C.; Guillaume, M.; Brohée, D.; Hubert, J.L.; Vanhaeverbeek, M. A new device for measurement of fibrin clot lysis: Application to the Euglobulin Clot Lysis Time. BMC Biotechnol. 2002, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Ilich, A.; Kumar, V.; Ferrara, M.J.; Henderson, M.W.; Noubouossie, D.F.; Jenkins, D.H.; Kozar, R.A.; Park, M.S.; Key, N.S. Euglobulin clot lysis time reveals a high frequency of fibrinolytic activation in trauma. Thromb. Res. 2021, 204, 22–28. [Google Scholar] [CrossRef]
- Smith, A.A.; Jacobson, L.J.; Miller, B.I.; Hathaway, W.E.; Manco-Johnson, M.J. A new euglobulin clot lysis assay for global fibrinolysis. Thromb. Res. 2004, 112, 329–337. [Google Scholar] [CrossRef]
- Schöchl, H.; Nienaber, U.; Hofer, G.; Voelckel, W.; Jambor, C.; Scharbert, G.; Kozek-Langenecker, S.; Solomon, C. Goal-directed coagulation management of major trauma patients using thromboestatometry (ROTEM)-guided administration of fibrinogen concentration and prothrombin complex concentrate. Crit. Care 2010, 14, R55. [Google Scholar] [CrossRef]
- Mou, Q.; Zhou, Q.; Liu, S. Blood clot parameters: Prejudgment of fibrinolysis in thromboelastography. Clin. Chim. Acta 2018, 479, 94–97. [Google Scholar] [CrossRef]
- Hett, D.A.; Walker, D.; Pilkington, S.N.; Smith, D.C. Sonoclot analysis. Br. J. Anaesth. 1995, 75, 771–776. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 2016, 6, 174. [Google Scholar] [CrossRef]
- Banerjee, A.; Chisti, Y.; Banerjee, U.C. Streptokinase—A clinically useful thrombolytic agent. Biotechnol. Adv. 2003, 22, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Collen, D.; Lijnen, H.R.; Vanderschueren, S. Staphylokinase: Fibrinolytic properties and current experience in patients with occlusive arterial thrombosis. Verh.-K. Acad. Voor Geneeskd. Van Belg. 1995, 57, 183–196. [Google Scholar]
- Choi, N.S.; Chang, K.T.; Jae Maeng, P.; Kim, S.H. Cloning, expression and fibrin(ogen)olytic properties of a Subtilisn DJ-4 Gene from Bacillus sp. DJ 4. FEMS Microbiol. Lett. 2004, 236, 325–331. [Google Scholar] [CrossRef]
- Sharkova, T.S.; Kornienko, E.I.; Osmolovskii, A.A.; Kreier, V.G.; Baranova, N.A.; Egorov, N.S. Morphological and physiological properties of the micromycete Arthrobotrys longa, a producer of longolytin, a proteolytic complex with a thrombolytic effect. Microbiology 2016, 85, 180–184. [Google Scholar] [CrossRef]
- Shirasaka, N.; Naitou, M.; Okamura, K.; Kusuda, M.; Fukuta, Y.; Terashita, T. Purification and characterization of a fibrinolytic protease from Aspergillus oryzae KSK-3. Mycoscience 2012, 53, 354–364. [Google Scholar] [CrossRef]
- Osmolovskiy, A.A.; Rukavitsyna, E.D.; Kreier, V.G.; Baranova, N.A.; Egorov, N.S. Production of proteinases with fibrinolytic and fibrinogenolytic activity by a micromycete Aspergillus ochraceus. Microbiology 2017, 86, 512–516. [Google Scholar] [CrossRef]
- Zhao, L.; Lin, X.; Fu, J.; Zhang, J.; Tang, W.; He, Z. A Novel Bi-Functional Fibrinolytic Enzyme with Anticoagulant and Thrombolytic Activities from a Marine-Derived Fungus Aspergillus versicolor ZLH-1. Mar. Drugs 2022, 20, 356. [Google Scholar] [CrossRef]
- Wu, B.; Wu, L.; Chen, D.; Yang, Z.; Luo, M. Purification and characterization of a novel fibrinolytic protease from Fusarium sp. CPCC 480097. J. Ind. Microbiol. Biotechnol. 2009, 36, 451–459. [Google Scholar] [CrossRef]
- Baggio, L.M.; Panagio, L.A.; Gasparin, F.G.M.; Sartori, D.; Celligoi, M.A.P.C.; Baldo, C. Production of fibrinogenolytic and fibrinolytic enzymes by a strain of Penicillium sp. Isolated from contaminated soil with industrial effluent. Acta Scientiarum. Health Sci. 2019, 41, e40606. [Google Scholar] [CrossRef]
- Liu, X.-L.; Du, L.-X.; Lu, F.-P.; Zheng, X.Q.; Xiao, J. Purification and characterization of a novel fibrinolytic enzyme from Rhizopus chinensis 12. Appl. Microbiol. Biotechnol. 2005, 67, 209–214. [Google Scholar]
- Kornienko, E.I.; Osmolovskiy, A.A.; Kreyer, V.G.; Baranova, N.A.; Kotova, I.B.; Egorov, N.S. Characteristics and Properties of the Complex of Proteolytic Enzymes of the Thrombolytic Action of the Micromycete Sarocladium strictum. Appl. Biochem. Microbiol. 2021, 57, 57–64. [Google Scholar] [CrossRef]
- Liu, X.; Kopparapu, N.K.; Li, Y.; Deng, Y.; Zheng, X. Biochemical characterization of a novel fibrinolytic enzyme from Cordeceps militaris. Int. J. Biol. Macromol. 2016, 94, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, J.S.; Kim, J.E.; Sapkota, K.; Shen, M.H.; Kim, S.; Chun, H.S.; Yoo, J.C.; Choi, H.S.; Kim, M.K.; et al. Purification and characterisation of fibrinolytic enzyme from cultured mycelia of Armillaria mella. Protein Expr. Purif. 2005, 43, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Mótyán, J.A.; Tóth, F.; Tőzsér, J. Research application in proteolytic enzymes in molecular microbiology. Biomolecules 2013, 3, 923–942. [Google Scholar] [CrossRef]
- Ryan, B.J.; Henehan, G.T. Overview of Approaches to Preventing and Avoiding Proteolysis During Expression and Purification of Proteins. Curr. Protoc. Protein Sci. 2013, 71, 5–25. [Google Scholar] [CrossRef]
- Hedstrom, L. Serine Protease mechanism and specificity. Chem. Rev. 2002, 102, 4501–4523. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J. Evolutionary families of peptidases. Biochem. J. 1993, 290, 205–218. [Google Scholar] [CrossRef]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and Biotechnological Aspects of Microbial Proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635. [Google Scholar] [CrossRef]
- Wu, J.W.; Chen, X.L. Extracellular metalloprotease from bacteria. Appl. Microbiol. Biotechnol. 2011, 92, 253–262. [Google Scholar] [CrossRef]
- Fukasawa, K.M.; Hata, T.; Ono, Y.; Hirose, J. Metal Preferences of Zinc-Binding Motif on Metalloproteases. J. Amino Acids 2011, 2011, 574816. [Google Scholar] [CrossRef]
- Ethiraj, S.; Gopinat, S. Production, purification, characterization, immobilization, and application of Serrapeptase: A review. Front. Biol. 2017, 12, 333–348. [Google Scholar] [CrossRef]
- Wu, S.; Feng, C.; Zhong, J.; Huan, L. Roles of S3 Site Residues of Nattokinase on Its Activity and Substrate Specificity. Biochem. J. 2007, 142, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.D.; Vaithilingam, M.; Shanker, R.; Kumar, S.; Thiyur, S.; Babu, V.; Selvakumar, J.N.; Prakash, S. Exploring the In Vitro Thrombolytic Activity of Nattokinase from a New Strain Pseudomonas aeruginosa CMSS. Jundishapur J. Microbiol. 2015, 8, e23567. [Google Scholar] [CrossRef]
- Kim, W.; Choi, K.; Kim, Y.; Park, H.; Choi, J.; Lee, Y.; Oh, H.; Kwon, I.; Lee, S. Purification and Characterization of a Fibrinolytic Enzyme Produced from Bacillus sp. Strain CK 11-4 Screened from Chungkook-Jang. Appl. Environ. Microbiol. 1996, 62, 2482–2488. [Google Scholar] [CrossRef] [PubMed]
- Yamane, T.A.; Kani, T.A.; Hatanaka, T.A.; Suzuki, A.T.; Ashida, T.A.; Kobayashi, T.; Ito, S.; Yamashita, O. Structure of a New Alkaline Serine Protease (M-Protease) from Bacillus sp. KSM-KI6. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 51, 199–206. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, G.T.; Kim, D.K.; Choi, W.A.; Park, S.H.; Jeong, Y.K.; Kong, I.S. Purification and characterisation of a novel fibrinolytic enzyme from Bacillus sp. KA 38 originated from fermented fish. J. Ferment. Bioeng. 1997, 84, 307–312. [Google Scholar]
- Devaraj, Y.; Rajender, S.K.; Halami, P.M. Purification, characterization of fibrinolytic protease from Bacillus amyloliquefaciens MCC2606 and analysis of fibrin degradation product by MS/MS. Prep. Biochem. Biotechnol. 2018, 48, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Agrebi, R.; Hmidet, N.; Hajji, M.; Ktari, N.; Haddar, A.; Fakhfakh-Zouari, N.; Nasri, M. Fibrinolytic Serine Protease Isolation from Bacillus amyloliquefaciens An6 Grown on Mirabilis jalapa Tuber Powders. Appl. Biochem. Biotechnol. 2010, 162, 75–88. [Google Scholar] [CrossRef]
- Ko, J.H.; Yan, J.P.; Zhu, L.; Qi, Y.P. Identification of two novel fibrinolytic enzymes from Bacillus subtilis QK02. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2003, 137, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, R.-H.; Peng, Y.; Zhang, Y.-Z. Highly efficient gene expression of a fibrinolytic enzyme (subtilisin DFE) in Bacillus subtilis mediated by the promoter of α-amylase gene from Bacillus amyloliquefaciens. Biotechnol. Lett. 2004, 26, 1365–1369. [Google Scholar] [CrossRef]
- Velusamy, P.; Pachaiappan, R.; Christopher, M.; Vaseeharan, B.; Anbu, P.; So, J.-S. Isolation and identification of a novel fibrinolytic Bacillus tequilensis CWD-67 from dumping soils enriched with poultry wastes. J. Gen. Appl. Microbiol. 2015, 61, 241–247. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yeo, W.S.; Seo, M.J.; Kim, M.J.; Lee, H.H.; Kang, B.W.; Park, J.U.; Choi, Y.H.; Jeong, Y.K. Biochemical analysis of fibrinolytic enzyme purified from Bacillus subtilis strain A1. J. Microbiol. 2011, 49, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Bae, D.H.; Kwon, T.J.; Lee, S.B.; Lee, H.H.; Park, J.H.; Heo, S.; Johnson, M.G. Purification and Characterization of a fibrinolytic enzyme from Bacillus sp. KDO-13 Isolated from Soyabean Paste. J. Microbiol. Biotechnol. 2001, 11, 845–852. [Google Scholar]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPS. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Arun, A.; Vincent, S.G.P.; Arasu, M.V.; Al-Dhabi, N.A. Cow Dung Is a Novel Feedstock for Fibrinolytic Enzyme Production from Newly Isolated Bacillus sp. IND 7 and Its Application in In Vitro Clot Lysis. Front. Microbiol. 2016, 7, 361. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Rai, S.K.; Thakur, R.; Chattopadhyay, P.; Kar, S.K. Bafibrinase: A non-toxic, non-hemorrhagic, direct-acting fibrinolytic serine protease from Bacillus sp. Strain AS-S20-I exhibits in vivo anticoagulant activity and thrombolytic potency. Biochimie 2012, 94, 1300–1308. [Google Scholar] [CrossRef]
- Jeong, Y.K.; Park, J.U.; Baek, H.; Park, S.H.; Kong, I.S.; Kim, D.W.; Joo, W.H. Purification and biochemical characterization of a fibrinolytic enzyme from Bacillus subtilis BK-17. World J. Microbiol. Biotechnol. 2001, 17, 89–92. [Google Scholar] [CrossRef]
- Paik, H.D.; Lee, S.K.; Heo, S.; Kim, S.Y.; Lee, H.H.; Kwon, T.J. Purification and Characterization of the Fibrinolytic Enzyme Produced by Bacillus subtilis KCK-7 from Chungkookjang. J. Microbiol. Biotechnol. 2004, 14, 829–835. [Google Scholar]
- Yuan, J.; Yang, J.; Zhuang, Z.; Yang, Y.; Lin, L.; Wang, S. Thrombolytic effects of Douchi Fibrinolytic enzyme from Bacillus subtilis LD-8547 in vitro and in vivo. BMC Biotechnol. 2012, 12, 36. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Prakash Vincent, S.G. Medium Optimization for the Production of Fibrinolytic Enzyme by Paenibacillus sp. IND8 Using Response Surface Methodology. Sci. World J. 2014, 2014, 276942. [Google Scholar] [CrossRef]
- Wang, J.; Wang, M.; Wang, Y. Purification and characterization of a novel fibrinolytic enzymes from Streptomyces sp. Clin. J. Biotechnol. 1999, 15, 83–89. [Google Scholar]
- Verma, P.; Chatterjee, S.; Keziah, M.S.; Devi, S.C. Fibrinolytic protease from marine Streptomyces rubiginosus VITPSSM. Cardiovasc. Hematol. Agents Med. Chem. 2018, 16, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Chitte, R.R.; Deshmukh, S.V.; Kanekar, P.P. Production, Purification, and Biochemical Characterization of a Fibrinolytic Enzyme from Thermophilic Streptomyces sp. MCMB-379. Appl. Biochem. Biotechnol. 2011, 165, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Babu, V.; Devi, C.S. Exploring the in vitro thrombolytic potential of streptokinase-producing β-hemolytic Streptococci isolated from bovine milk. J. Gen. Appl. Microbiol. 2015, 61, 139–146. [Google Scholar] [CrossRef]
- Narasimhan, M.K.; Ethiraj, S.; Krishnamurthi, T.; Rajesh, M. Purification, biochemical and thermal properties of fibrinolytic enzyme secreted by Bacillus cereus SRM-001. Prep. Biochem. Biotechnol. 2017, 48, 34–42. [Google Scholar] [CrossRef]
- Biji, G.D.; Arun, A.; Muthulakshmi, E.; Vijayaraghavan, P.; Arasu, M.V.; Al-Dhabi, N.A. Bio-prospecting of cuttle fish waste and cow dung for the production of fibrinolytic enzyme from Bacillus cereus IND5 in solid state fermentation. 3 Biotech 2016, 6, 231. [Google Scholar] [CrossRef]
- Afifah, D.N.; Sulchan, M.; Syah, D.; Yanti, Y.; Suhartono, M.T.; Kim, J.H. Purification and Characterization of a Fibrinolytic Enzyme from Bacillus pumilus 2.g Isolated from Gembus, an Indonesian Fermented Food. Prev. Nutr. Food Sci. 2014, 19, 213–219. [Google Scholar] [CrossRef]
- Taneja, K.; Bajaj, B.K.; Kumar, S.; Dilbaghi, N. Production, purification and characterization of fibrinolytic enzyme from Serratia sp. KG-2-1 using optimized media. 3 Biotech 2017, 7, 184. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Prakash Vincent, S.G. A low-cost fermentation medium for potential fibrinolytic enzyme production by a newly isolated marine bacterium Shewanella sp. IND 20. Biotechnol. Rep. 2015, 7, 135–142. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meshram, V.; Saxena, S. Potential fibrinolytic activity of an endophytic Lasiodiplodia pseudotheobromae species. 3 Biotech 2016, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, T.P.; Sales, A.E.; Porto, T.S.; Costa, R.M.P.B.; Breydo, L.; Uversky, V.N.; Porto, A.L.F.; Converti, A. Purification, biochemical, and structural characterization of a novel fibrinolytic enzyme from Mucor subtilissimus UCP 1262. Bioprocess Biosyst. Eng. 2017, 40, 1209–1219. [Google Scholar] [CrossRef]
- Abdel-Fattah, A.F.; Ismail, A.M.S. Preparation and Properties of Fibrinolytic Enzymes Produced by Cochliobolus lunatus. Biotechnol. Bioeng. 1983, 26, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Yao, J.; Sparks, S.; Wang, K.Y. Nattokinase: An Oral Antithrombotic Agent for the Prevention of Cardiovascular Disease. Int. J. Mol. Sci. 2017, 18, 523. [Google Scholar] [CrossRef]
- Meruvu, H.; Vangalapati, M. Nattokinase: A Review on Fibrinolytic Enzyme. Int. J. Chem. Environ. Pharm. Res. 2011, 2, 61–66. [Google Scholar]
- RCSB PDB. Available online: http://www.rcsb.org/structure/4DWW (accessed on 15 September 2023).
- Cal, D.; Zhu, C.; Chen, S. Microbial production of nattokinase: Current, challenge and prospect. World J. Microbiol. Biotechnol. 2017, 33, 84. [Google Scholar]
- Yongjun, C.; Wei, B.; Shujun, J.; Meizhi, W.; Yan, J.; Yan, Y.; Zhongliang, Z.; Goulin, Z. Directed evolution improves the fibrinolytic activity of nattokinase from Bacillus natto. FEMS Microbiol. Lett. 2011, 325, 155–161. [Google Scholar] [CrossRef]
- Sikri, N.; Bardia, A. A History of Streptokinase use in Acute Myocardial Infarction. Tex. Heart Inst. J. 2007, 34, 318–327. [Google Scholar]
- RCSB PDB. Available online: https://www.rcsb.org/structure/1L4D (accessed on 15 September 2023).
- Vakili, B.; Nezafat, N.; Negahdaripour, M.; Yari, M.; Zare, B.; Ghasemi, Y. Staphylokinase Enzyme: An Overview of Structure, Function and Engineered Forms. Curr. Pharm. Biotechnol. 2017, 18, 1026–1037. [Google Scholar] [CrossRef]
- Peetermans, M.; Vanassche, T.; Liesenborghs, L.; Claes, J.; Velde, G.V.; Kwiecinksi, J.; Jin, T.; De Geest, B.; Hoylaerts, M.F.; Lijnen, R.H.; et al. Plasminogen activation by staphylokinase enhances local spreading of S. aureus in skin infections. BMC Microbiol. 2014, 14, 310. [Google Scholar] [CrossRef]
- RCSB PDB. Available online: http://www.rcsb.org/structure/1C78 (accessed on 15 September 2023).
- Nguyen, L.T.; Vogel, H.J. Staphylokinase has distinct modes of interaction with antimicrobial peptides, modulating its plasminogen-activation properties. Sci. Rep. 2016, 6, 31817. [Google Scholar] [CrossRef]
- RCSB PDB. Available online: https://www.rcsb.org/structure/1SRP (accessed on 15 September 2023).
- Selan, L.; Papa, R.; Tilotta, M.; Vrenna, G.; Carpentieri, A.; Amoresano, A.; Pucci, P.; Artini, M. Serratiopeptidase: A well-known metalloprotease with a new non-proteolytic activity against S. aureus biofilm. BMC Microbiol. 2015, 15, 207. [Google Scholar] [CrossRef]
- Gupte, V.; Luthra, U. Analytical techniques for Serratiopeptidase: A review. J. Pharm. Anal. 2017, 7, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Nirale, N.M.; Menon, M.D. Topical Formulations of Serratiopeptidase: Development and Pharmacodynamic Evaluation. Indian J. Pharm. Sci. 2010, 72, 65–71. [Google Scholar] [PubMed]
- Podorol’skaya, L.V.; Serebryakova, T.N.; Sharkova, T.S.; Neumyvakin, L.V.; Khromov, I.S.; Khokhlov, N.V.; Tarantul, V.Z. Tarantul Experimental thrombosis in rabbit marginal ear vein and evaluation of the thrombolytic action of longolytin. Bull. Exp. Biol. Med. 2007, 143, 577–580. [Google Scholar] [CrossRef]
- Sharkova, T.; Podorolskaya, L. Increase of Fibrinilytic and Anticoagulant Activity by per os Administrating of Proteinase Complex Longolytin in Rats. J. Thromb. Haemost. 2017, 16, 355. [Google Scholar]
- Osmolovskiy, A.A.; Kreyer, V.G.; Baranova, N.A.; Egorov, N.S. Proteolytic Enzymes of Mycelial Fungi with Plasmin-like and Plasminogen-Activator Activity. Usp. Sovr. Biol. 2021, 141, 467–482. [Google Scholar]
- Kurosawa, Y.; Nirengi, S.; Homma, T.; Esaki, K.; Ohta, M.; Clark, J.F.; Hamaoka, T. A single-dose of oral nattokinase potentiates thrombolysis and anti-coagulation profiles. Sci. Rep. 2015, 5, 11601. [Google Scholar] [CrossRef] [PubMed]
- Ly, B.; Arnesen, H.; Eie, H.; Hol, R. A controlled clinical trial of streptokinase and heparin in the treatment of major pulmonary embolism. Acta Med. Scand. 1978, 203, 465–470. [Google Scholar] [CrossRef]
- Gusev, E.I.; Martynov, M.Y.; Nikonov, A.A.; Shamalov, N.A.; Semenov, M.P.; Gerasimets, E.A.; Yarovaya, E.B.; Semenov, A.M.; Archakov, A.I.; Markin, S.S.; et al. Non-immunogenic recombinant staphylokinase versus alteplase for patients with acute ischaemic stroke 4.5 h after symptom onset in Russia (FRIDA): A randomised, open label, multicentre, parallel-group, non-inferiority trial. Lancet Neurol. 2021, 20, 721–728. [Google Scholar] [CrossRef]
- Kim, J.Y.; Gum, S.N.; Paik, J.K.; Lim, H.H.; Kim, K.C.; Ogasawara, K.; Inoue, K.; Park, S.; Jang, Y.; Lee, J.H. Effects of nattokinase on blood pressure: A randomized, controlled trial. Hypertens. Res. 2008, 31, 1583–1588. [Google Scholar] [CrossRef]
- Fadl, N.N.; Ahmed, H.H.; Booles, H.F.; Sayed, A.H. Serrapeptase and nattokinase intervention for relieving Alzheimer’s disease pathophysiology in rat model. Hum. Exp. Toxicol. 2013, 32, 721–735. [Google Scholar] [CrossRef]
- Moriya, N.; Nakata, M.; Nakamura, M.; Takaoka, M.; Iwasa, S.; Kato, K.; Kakinuma, A. Intestinal absorption of serrapeptase (TSP) in rats. Biotechnol. Appl. Biochem. 1994, 20, 101–108. [Google Scholar] [CrossRef]
- Kotb, E. Fibrinolytic bacterial enzymes with thrombolytic activity. In Fibrinolytic Bacterial Enzymes with Thrombolytic Activity; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Majima, Y.; Inagaki, M.; Hirata, K.; Takeuchi, K.; Morishita, A.; Sakakura, Y. The effect of an orally administered proteolytic enzyme on the elasticity and viscosity of nasal mucus. Arch. Otorhinolaryngol. 1988, 44, 355–359. [Google Scholar] [CrossRef]
- Bracale, G.; Selvetella, L. Clinical study of the efficacy of and tolerance to seaprose S in inflammatory venous disease. Controlled study versus serratio-peptidase. Minerva Cardioangiol. 1996, 44, 515–524. [Google Scholar] [PubMed]
- Esch, P.M.; Gerngross, H.; Fabian, A. Reduction of postoperative swelling. Objective measurement of swelling of the upper ankle joint in treatment with serrapeptase: A prospective study. Fortschritte Der Med. 1989, 107, 67–68. [Google Scholar]
- Mazzone, A.; Catalani, M.; Costanzo, M.; Drusian, A.; Mandoli, A.; Russo, S.; Guarini, E.; Vesperini, G. Evaluation of Serratia peptidase in acute or chronic inflammation of otorhinolaryngology pathology: A multicentre, double blind, randomized trial versus placebo. J. Int. Med. Res. 1990, 18, 379–388. [Google Scholar] [CrossRef]
- Mine, Y.; Wong, A.H.K.; Jiang, B. Fibrinolytic enzymes in Asian traditional fermented foods. Food Res. Int. 2005, 38, 243–250. [Google Scholar] [CrossRef]
- Wong, A.H.; Mine, Y. Novel fibrinolytic enzyme in fermented shrimp paste, a traditional Asian fermented seasoning. J. Agric. Food Chem. 2004, 52, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Quyen, T.D.; Le, H.T. Cloning and enhancing production of a detergent- and organic-solvent-resistant nattokinase from Bacillus subtilis VTCC-DVN-12-01 by using an eight-protease-gene-deficient Bacillus subtilis WB800. Microb. Cell Factories 2013, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, P.; Rajendran, P.; Prakash Vincent, S.G.; Arun, A.; Abdullah Al-Dhabi, N.; Valan Arasu, M.; Young Kwon, O.; Kim, Y.O. Novel sequential screening and enhanced production of fibrinolytic enzyme by Bacillus sp. IND12 using response surface methodology in solid-state fermentation. BioMed Res. Int. 2017, 2017, 3909657. [Google Scholar] [CrossRef] [PubMed]



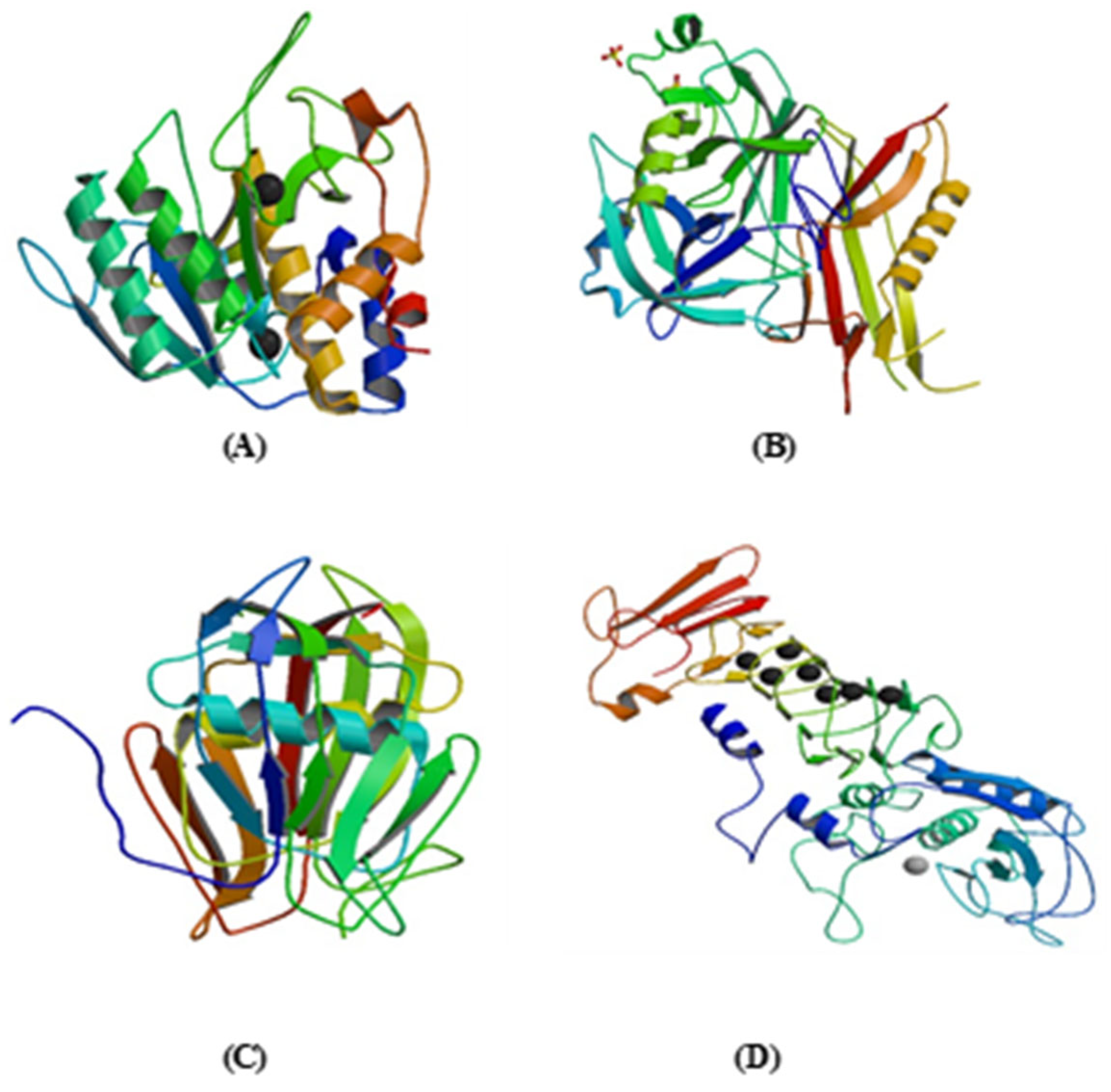
| Fibrinolytic Enzymes | Micro-Organisms Associated | Sources for Production | Physicochemical Properties | Functional Moiety | Mechanism of Action | References |
|---|---|---|---|---|---|---|
| Streptokinase | Streptococcus hemolyticus | Exudates of infected wounds | 47 kDa pH 7.5 37 °C | Single polypeptide chain (414 amino acids) with multiple structural domains (α, β, Υ) | plasminogen activation for the formation of β-domain SK plasminogen complex | [75,93] |
| Staphylokinase | Staphylococcus aureus | Human skin | 15.5 kDa pH 8.5 37 °C | Single polypeptide chain (136 amino acids) without disulphide bridge | plasminogen activation due to higher affinity with plasmin | [76,94] |
| Serrapeptase | Serratia marcescens E 15 | Intestine of silkworm | 45–60 kDa pH 9.0 40 °C | Metalloprotease with one active site and three Zn atoms | Cleavage of peptide bond linkages | [95] |
| Nattokinase (wild Type) | Bacillus subtilis YF 38, natto | Fermented soybean Natto | 27.7 kDa pH 8.6 | Presence of catalytic triad (Asp-32, His-64 and Ser-221) and one oxyanion hole (Asn-155) | Resemblance with plasmin and enhanced production of plasmin and clot dissolving agents | [8,96] |
| Nattokinase | Pseudomonas aeruginosa CMSS (new strain) | Cow milk | 21 kDa pH 7.0 25 °C | Like wild-type nattokinase with a two-fold increase in enzyme activation | Same as wild type nattokinase | [97] |
| CK fibrinolytic enzyme | Bacillus sp. CK | Chungkook-jang (Korea) | 28.2 kDa pH 10.5 70 °C | Thermolytic alkaline serine protease (1882 protein atoms, 2 Ca2+ ions, and 44 water molecules) | Higher tissue plasminogen activator production. | [98,99] |
| Fibrinolytic enzyme | Bacillus sp. KA38 | Jeot-gal (fermented fish, Korea) | 41 kDa pH 7.0 40 °C | Metalloprotease | Degrade fibrin or form plasmin from plasminogen | [100] |
| CFR 15 protease | Bacillus amyloliquefaciens MCC2606 (strain CFR 15) | Dosa batter | 32 kDa pH 10.5 45 °C | Serine protease with a catalytic triad (His-57, Ser-195, Asp-102) | Hydrolysis of αα-, ββ-, ϓ- chains of fibrin | [101] |
| B. amyloliquefaciens An6 fibrinase (BAF1) | Bacillus amyloliquefaciens An6 | Mirabilis jalapa tuber powder (MJTP) | 30 kDa pH 9.0 60 °C | Serine protease | Degrade fibrin or form plasmin from plasminogen | [102] |
| Subtilisin DJ-4 | Bacillus sp. DJ -4 | Doen-jang, Korea | 29 kDa pH 10.0 40 °C | Plasmin-like serine protease | Rapid hydrolysis of αα-, ββ-, ϓ- chains of fibrin | [77] |
| Subtilisin QK02 | Bacillus sp. QK02 | Fermented soybean | 28 kDa pH 8.5 55 °C | Serine protease with a catalytic triad (Asp-32, His-64 and Ser-221) | Cleaves peptide bond linkages | [103] |
| Subtilisin DFE | Bacillus amyloliquefaciens DC 4 | Douchi (China) | 28 kDa pH 9.0 48 °C | Serine protease | High specificity towards fibrin and hydrolyses thrombin | [104] |
| Fibrinolytic enzyme | Bacillus tequilensis CWD-67 | Dumping soil | 22 kDa pH 8.0 45 °C | Chymotrypsin-like serine metalloprotease containing hydrophobic S1 pocket | Hydrolysis of αα-, ββ-, ϓ- chains of fibrin | [105] |
| BacillokinaseII | Bacillus subtilis A1 | Local soil (Korea) | 31.4 kDa pH 7.0 50 °C | Chymotrypsin-like serine protease | Degrade fibrin and act as plasminogen activator | [106] |
| Fibrinolytic enzyme | Bacillus sp. KDO- 13 | Soybean paste (Korea) | 45 kDa pH 7 60 °C | Metalloprotease with Catalytic domain with 170 amino acids, hinge region, and hemopexin domain of 200 amino acids | Degrade fibrin or form plasmin from plasminogen | [107,108] |
| Fibrinolytic enzyme | Bacillus thuringiensis IND 7 | Cow dung | 32 kDa pH 9.0 | Serine protease | Degrade fibrin or form plasmin from plasminogen | [109] |
| Bafibrinase | Bacillus Sp. AS-S20-I | Soil (Assam) | 32.3 kDa 7.4 pH 37 °C | Catalytic triad (Ser-221, His-64 and Asp-32) without intramolecular sulphide bond | Cleaves chains of fibrin (α, β) and fibrinogen | [110] |
| Subtilisin BK 17 | Bacillus subtilis BK17 | Decaying rice plant (Korea) | 31 kDa | Serine protease | Degrade fibrin or form plasmin from plasminogen | [111] |
| Fibrinolytic enzyme | Bacillus subtilis KCK-7 | Chungkookjang (fermented food) | 45 kDa pH 7.0 60 °C | Serine protease requires hydroxyl group for activity | Degrade fibrin or form plasmin from plasminogen | [112] |
| Douchi fibrinolytic enzyme | Bacillus subtilis LD 8547 | Soybean fermented food (China) | 30 kDa | Serine protease | Activate t-PA | [113] |
| Fibrinolytic enzyme | Paenibacillus sp. IND8 | Cooked Indian rice | - | - | Degrade fibrin or form plasmin from plasminogen | [114] |
| SW 1 | Streptomyces sp. Y405 | Soil isolate | 30 kDa pH 8.0 | Serine protease and metalloprotease | Degrade fibrin or form plasmin from plasminogen | [115] |
| Fibrinolytic enzyme | Streptomyces rubiginosus | Marine soil | 45 kDa pH 7.2 32 °C | - | Degrade fibrin or form plasmin from plasminogen | [116] |
| Fibrinolytic enzyme | Streptomyces sp. MCMB-379 | Seed culture | - | Serine endopeptidase type | Cleaves fibrin fibres by degradation of chains | [117] |
| β Haemolytic Streptokinase | Streptococcus equinus | Bovine milk | - | - | Degrade fibrin or form plasmin from plasminogen | [118] |
| Fibrinolytic enzyme | Bacillus cereus SRM-001 | Chicken dump yard | 28 kDa pH 7.0 37 °C | Serine protease | Plasmin catalysed hydrolysis of fibrin | [119] |
| Fibrinolytic enzyme | Bacillus cereus IND 5 | Cuttle fish waste and cow dung | 47 kDa pH 8.0 50 °C | Serine protease | Degrade fibrin or form plasmin from plasminogen | [120] |
| Fibrinolytic enzyme | Bacillus pumilus | Gembus (Indonesia fermented food) | 20 kDa 50 °C | Serine protease | Degrade α- and β-chains of fibrinogen but not Υ-chain | [121] |
| Fibrinolytic enzyme | Serratia sp. KG 2–1 | Garbage dump yard | pH 8.0 40 °C | Metalloprotease | Degrade fibrin or form plasmin from plasminogen | [122] |
| Fibrinolytic enzyme | Shewanella sp. IND20 | Fish Sardinella longiceps | 55.5 kDa pH 8.0 50 °C | Serine protease | Direct clot lysis and plasminogen activation activity | [123] |
| Fibrinolytic enzyme | Cordyceps militaris | Mushroom | 28 kDa pH 7.2 37 °C | Serine protease | Activate plasminogen to plasmin | [78] |
| Fibrinolytic enzyme | Lasiodiplodia pseudotheobromae | Aegle Marmelos (Golden apple) | 80 kDa | - | Degrade fibrin or form plasmin from plasminogen | [124] |
| AMMP | Armillaria mella | Mushroom (Korea) | 21 kDa pH 6.0 33 °C | Chymotrypsin like metalloprotease | Hydrolyse α-α fibrinogen | [79] |
| Fibrinolytic enzyme | Mucor subtilissimus UCP 1262 | Soil (Brazil) | 20 kDa pH 9.0 40 °C | Chymotrypsin like serine protease | Properties resemble to plasmin | [125] |
| Fibrinolytic enzyme | Cochliobolus lunatus | Surface culture | pH 6.8 40 °C | - | Degrade fibrin or form plasmin from plasminogen | [126] |
| Longolytin | Arthrobotrys longa | Soil, contains nematodes | 28.6 kDa pH 6.0–9.0 | Serine protease contains thiol groups | Hydrolyse fibrin and activate plasminogen like urokinase | [127,128] |
| Fibrinolytic enzyme | Aspergillus ochraceus L-1 | Soil | 36 kDa pH 10.0–11.0 45 °C | Serine protease | Hydrolyse fibrin and fibrinogen | [79] |
| Fibrinolytic enzyme | Aspergillus oryzae KSK-3 | Commercial rice-koji for miso brewing | 30 kDa pH 6.0 50 °C | Serine protease | Hydrolyse fibrin and fibrinogen | [80] |
| Versiase | Aspergillus versicolor | Marine sponge Callyspongia sp. | 37.3 kDa pH 5.0 40 °C | Metalloprotease | Hydrolyse fibrin directly and indirectly via the activation of plasminogen, and it can hydrolyse α-, β- and γ-chains of fibrinogen. | [81] |
| Fu-P | Fusarium sp. CPCC480097 | Shanghai Health Creation Center of Biopharmaceutical R&D | 28 kDa pH 8.5 45 °C | Serine protease | Hydrolyse fibrin and fibrinogen | [82] |
| Fibrinolytic enzyme | Paecilomyces tenuipes | Culture Collection of DNA Bank of Mushrooms, Incheon, Republic of Korea. | 14 kDa pH 5.0 35 °C | Serine protease | Hydrolyse the Aα chain of human fibrinogen, but do not hydrolyse the Bβ or γ chains | [83] |
| Fibrinolytic enzyme | Rhizopus chinensis 12 | Brewing rice wine | 18.0 kDa pH 10.5 45 °C | Serine protease. The first 12 amino acids of the N-terminal sequence of the enzyme were S-V-S-E-I-Q-L-M-H-N-L-G and had no homology with that of other fibrinolytic enzyme from other microorganism. | Hydrolyse fibrin and α-, β- and γ-chains of fibrinogen | [84] |
| Fibrinolytic enzyme | Sarocladium strictum 1 | Arhtrobotrys longa co-culture | 35.0 kDa pH 9.0 37 °C | Serine protease | Hydrolyse fibrin and activate plasminogen like urokinase | [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.; Gautam, P.; Sharma, C.; Osmolovskiy, A. Fibrin and Fibrinolytic Enzyme Cascade in Thrombosis: Unravelling the Role. Life 2023, 13, 2196. https://doi.org/10.3390/life13112196
Singh R, Gautam P, Sharma C, Osmolovskiy A. Fibrin and Fibrinolytic Enzyme Cascade in Thrombosis: Unravelling the Role. Life. 2023; 13(11):2196. https://doi.org/10.3390/life13112196
Chicago/Turabian StyleSingh, Rajni, Prerna Gautam, Chhavi Sharma, and Alexander Osmolovskiy. 2023. "Fibrin and Fibrinolytic Enzyme Cascade in Thrombosis: Unravelling the Role" Life 13, no. 11: 2196. https://doi.org/10.3390/life13112196
APA StyleSingh, R., Gautam, P., Sharma, C., & Osmolovskiy, A. (2023). Fibrin and Fibrinolytic Enzyme Cascade in Thrombosis: Unravelling the Role. Life, 13(11), 2196. https://doi.org/10.3390/life13112196








