Role of Ectopic Olfactory Receptors in the Regulation of the Cardiovascular–Kidney–Metabolic Axis
Abstract
:1. Introduction
2. Characterization of Mammalian Olfactory Receptors
2.1. Discovery of Olfactory Receptors in the Nasal Epithelium
2.2. Deorphanization of Olfactory Receptors
2.3. Olfactory Receptor Signaling Pathways
2.4. Classification and Nomenclature of Olfactory Receptors in Vertebrates
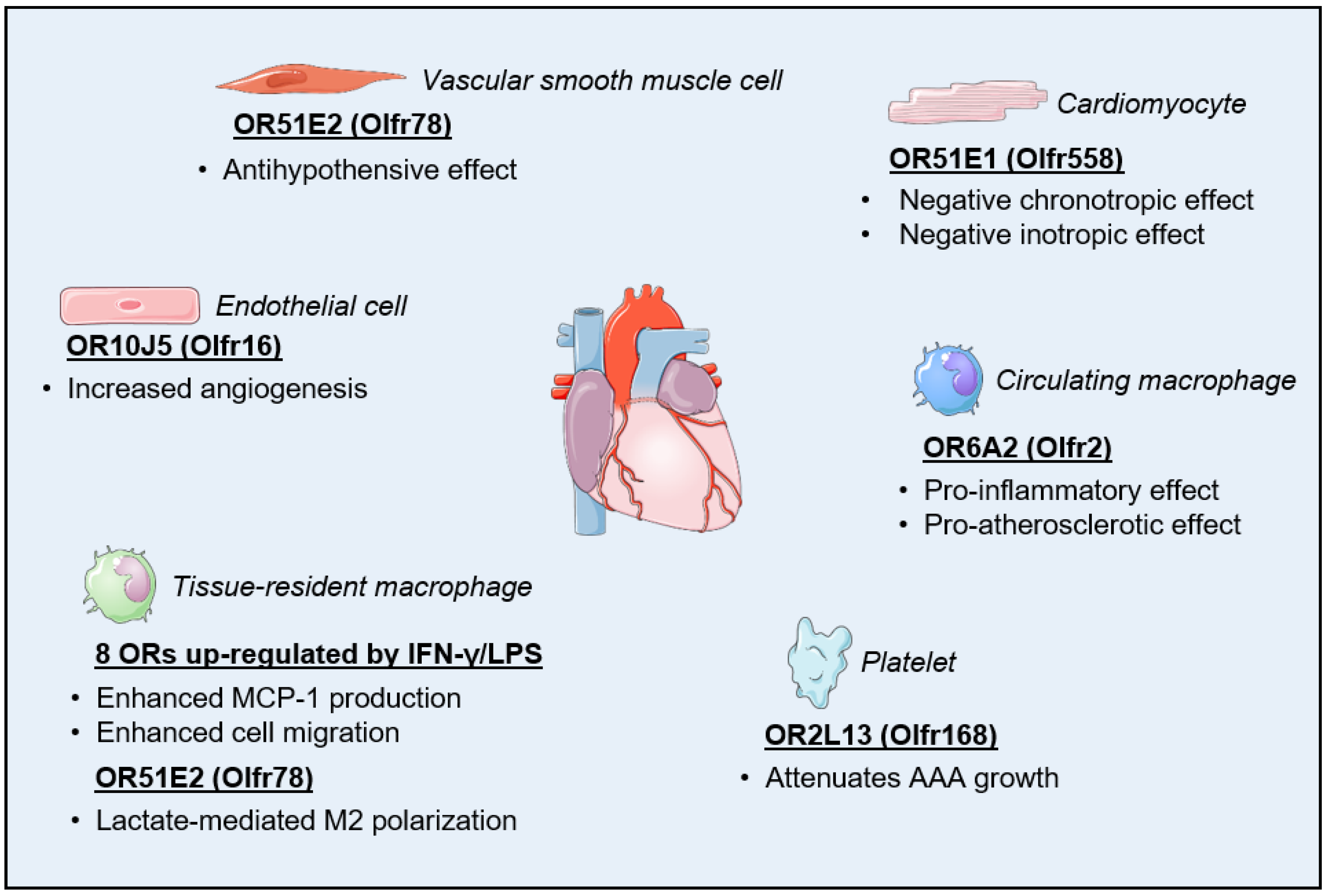
3. Olfactory Receptors in the Cardiovascular System
3.1. Regulation of Cardiac Function
3.2. Regulation of Vascular Function
3.3. Regulation of Inflammatory Processes
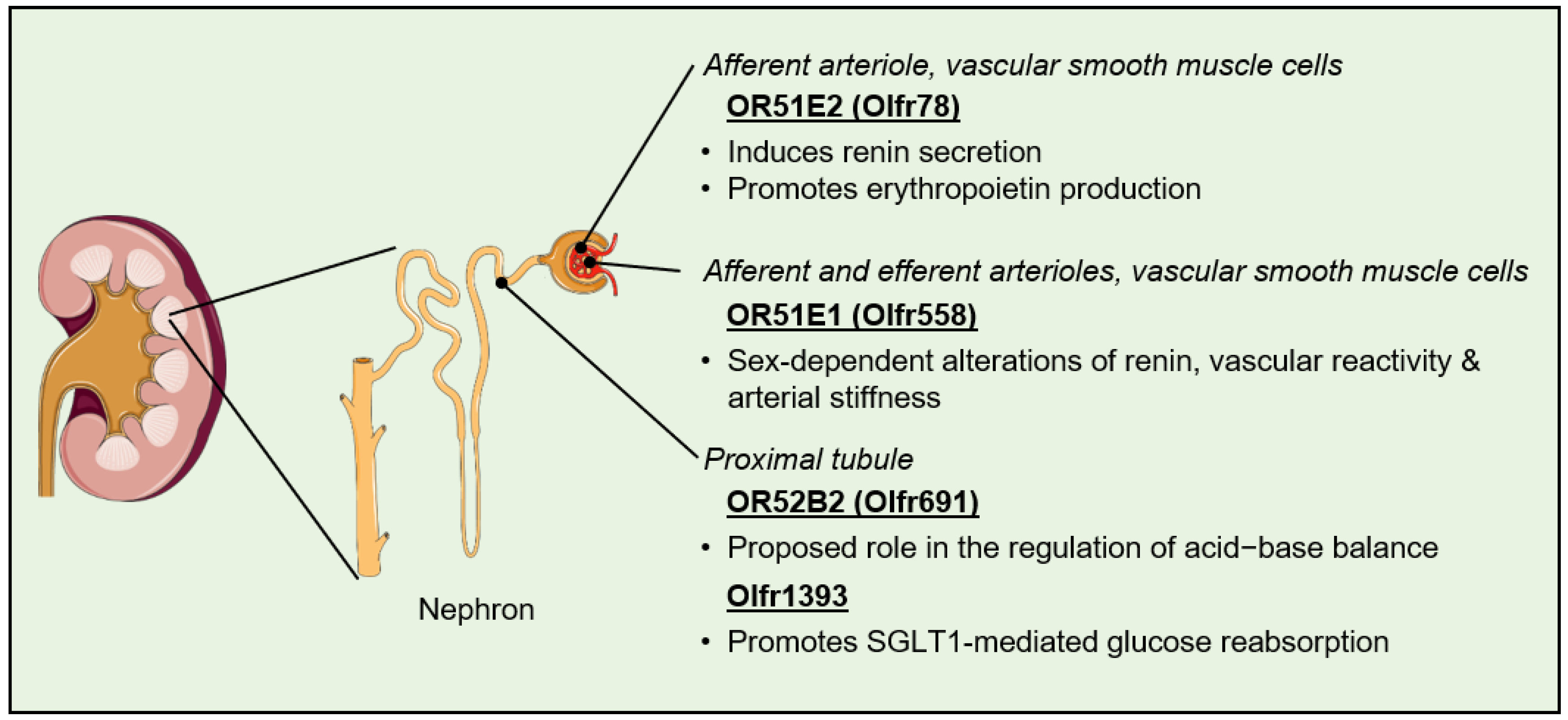
4. Olfactory Receptors in the Kidney
4.1. Regulation of Blood Pressure
4.2. Regulation of Fibrosis and Kidney Stone Formation
4.3. Regulation of Filtration, Reabsorption, and Secretion
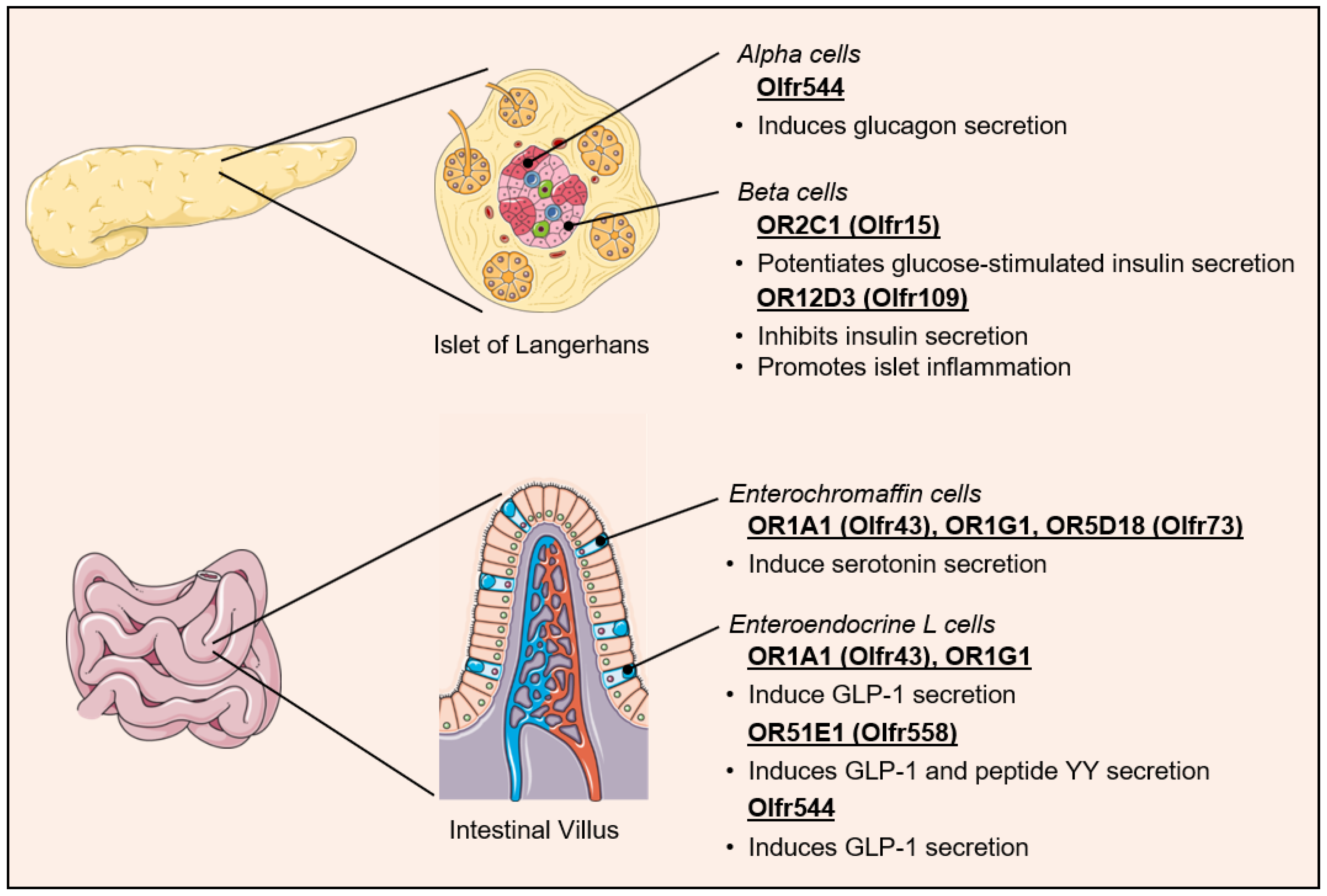
5. Ectopic Olfactory Receptors in the Control of Metabolic Homeostasis
5.1. Regulation of Pancreatic Hormone Secretion
5.2. Regulation of Incretin Secretion
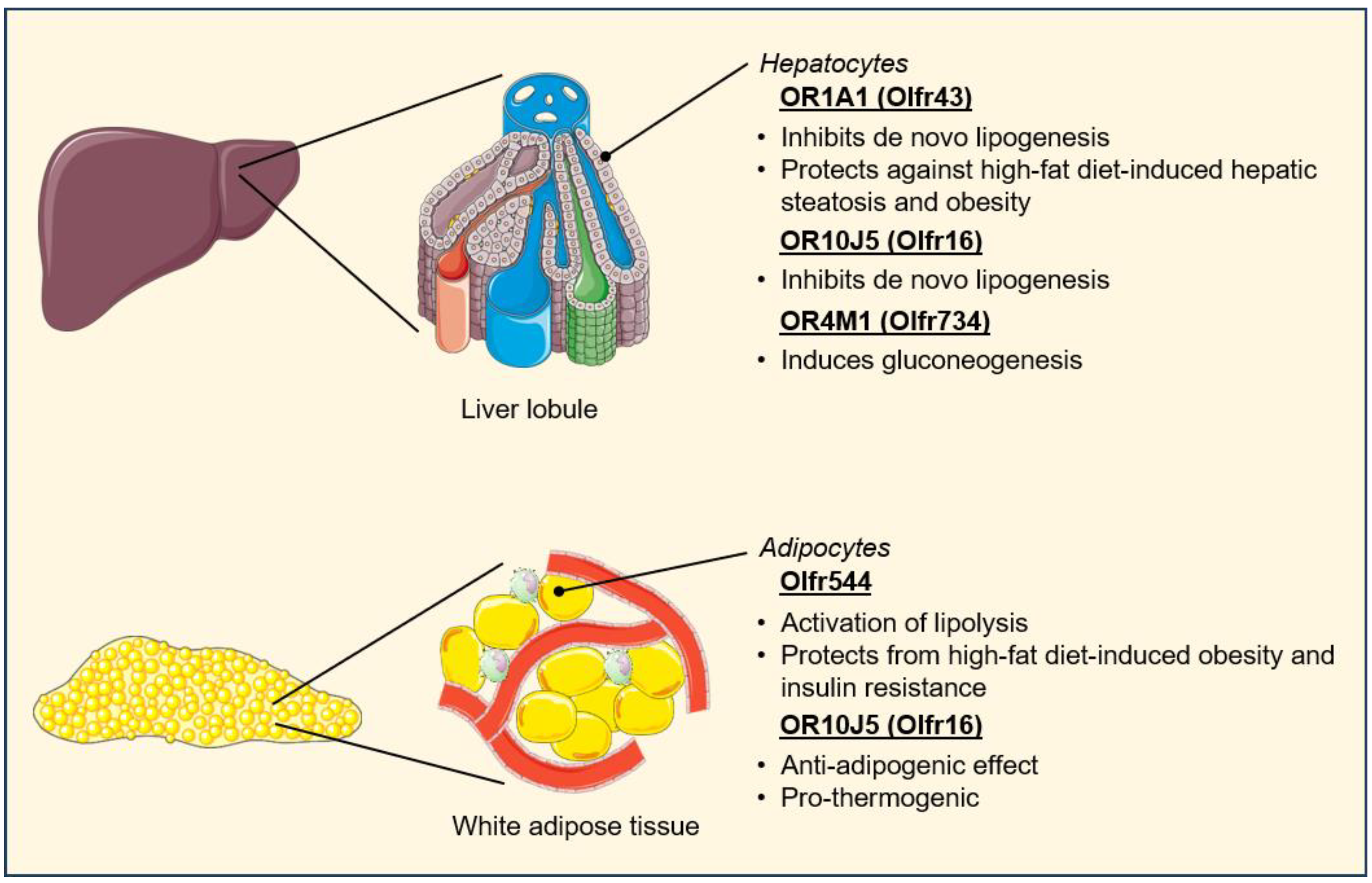
5.3. Regulation of Metabolic Tissue Function
5.4. Regulation of Cardiovascular–Renal Metabolic Health
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Addis, P.; Bali, U.; Baron, F.; Campbell, A.; Harborne, S.; Jagger, L.; Milne, G.; Pearce, M.; Rosethorne, E.M.; Satchell, R.; et al. Key aspects of modern GPCR drug discovery. SLAS Discov. 2024, 29, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Alhosaini, K.; Azhar, A.; Alonazi, A.; Al-Zoghaibi, F. GPCRs: The most promiscuous druggable receptor of the mankind. Saudi Pharm. J. 2021, 29, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhou, Q.; Labroska, V.; Qin, S.; Darbalaei, S.; Wu, Y.; Yuliantie, E.; Xie, L.; Tao, H.; Cheng, J.; et al. G protein-coupled receptors: Structure- and function-based drug discovery. Signal Transduct. Target. Ther. 2021, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gareri, C.; Rockman, H.A. G-Protein-Coupled Receptors in Heart Disease. Circ. Res. 2018, 123, 716–735. [Google Scholar] [CrossRef] [PubMed]
- Capote, L.A.; Mendez Perez, R.; Lymperopoulos, A. GPCR signaling and cardiac function. Eur. J. Pharmacol. 2015, 763, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Thai, B.S.; Chia, L.Y.; Nguyen, A.T.N.; Qin, C.; Ritchie, R.H.; Hutchinson, D.S.; Kompa, A.; White, P.J.; May, L.T. Targeting G protein-coupled receptors for heart failure treatment. Br. J. Pharmacol. 2023; ahead of print. [Google Scholar] [CrossRef]
- Leon, B.M.; Maddox, T.M. Diabetes and cardiovascular disease: Epidemiology, biological mechanisms, treatment recommendations and future research. World J. Diabetes 2015, 6, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Powell-Wiley, T.M.; Poirier, P.; Burke, L.E.; Despres, J.P.; Gordon-Larsen, P.; Lavie, C.J.; Lear, S.A.; Ndumele, C.E.; Neeland, I.J.; Sanders, P.; et al. Obesity and Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation 2021, 143, e984–e1010. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Ndumele, C.E.; Neeland, I.J.; Tuttle, K.R.; Chow, S.L.; Mathew, R.O.; Khan, S.S.; Coresh, J.; Baker-Smith, C.M.; Carnethon, M.R.; Despres, J.P.; et al. A Synopsis of the Evidence for the Science and Clinical Management of Cardiovascular-Kidney-Metabolic (CKM) Syndrome: A Scientific Statement From the American Heart Association. Circulation 2023, 148, 1636–1664. [Google Scholar] [CrossRef] [PubMed]
- Ndumele, C.E.; Rangaswami, J.; Chow, S.L.; Neeland, I.J.; Tuttle, K.R.; Khan, S.S.; Coresh, J.; Mathew, R.O.; Baker-Smith, C.M.; Carnethon, M.R.; et al. Cardiovascular-Kidney-Metabolic Health: A Presidential Advisory from the American Heart Association. Circulation 2023, 148, 1606–1635. [Google Scholar] [CrossRef] [PubMed]
- Bjarnadottir, T.K.; Gloriam, D.E.; Hellstrand, S.H.; Kristiansson, H.; Fredriksson, R.; Schioth, H.B. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics 2006, 88, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Insel, P.A.; Snead, A.; Murray, F.; Zhang, L.; Yokouchi, H.; Katakia, T.; Kwon, O.; Dimucci, D.; Wilderman, A. GPCR expression in tissues and cells: Are the optimal receptors being used as drug targets? Br. J. Pharmacol. 2012, 165, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Vassilatis, D.K.; Hohmann, J.G.; Zeng, H.; Li, F.; Ranchalis, J.E.; Mortrud, M.T.; Brown, A.; Rodriguez, S.S.; Weller, J.R.; Wright, A.C.; et al. The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 2003, 100, 4903–4908. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, J.; Breer, H.; Strotmann, J. Mammalian olfactory receptors. Front. Cell. Neurosci. 2009, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Drew, L. Olfactory receptors are not unique to the nose. Nature 2022, 606, S14–S17. [Google Scholar] [CrossRef] [PubMed]
- Massberg, D.; Hatt, H. Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Probst, W.C.; Snyder, L.A.; Schuster, D.I.; Brosius, J.; Sealfon, S.C. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 1992, 11, 1–20. [Google Scholar] [CrossRef]
- Firestein, S. How the olfactory system makes sense of scents. Nature 2001, 413, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Pilpel, Y.; Lancet, D. The variable and conserved interfaces of modeled olfactory receptor proteins. Protein Sci. 1999, 8, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Furudono, Y.; Sone, Y.; Takizawa, K.; Hirono, J.; Sato, T. Relationship between peripheral receptor code and perceived odor quality. Chem. Senses 2009, 34, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Martinez, Q.; Amson, E.; Laska, M. Does the number of functional olfactory receptor genes predict olfactory sensitivity and discrimination performance in mammals? J. Evol. Biol. 2024, 37, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Niimura, Y.; Nei, M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 2006, 51, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Haddad, R.; Khan, R.; Takahashi, Y.K.; Mori, K.; Harel, D.; Sobel, N. A metric for odorant comparison. Nat. Methods 2008, 5, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Harini, K.; Sowdhamini, R. Computational Approaches for Decoding Select Odorant-Olfactory Receptor Interactions Using Mini-Virtual Screening. PLoS ONE 2015, 10, e0131077. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Kubota, M.; Roberts, R.W.; Chi, Q.; Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell 2004, 119, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Cho, I.T.; Schoel, L.J.; Cho, G.; Golden, J.A. Hereditary spastic paraplegia-linked REEP1 modulates endoplasmic reticulum/mitochondria contacts. Ann. Neurol. 2015, 78, 679–696. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Matsunami, H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc. 2008, 3, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Bavan, S.; Sherman, B.; Luetje, C.W.; Abaffy, T. Discovery of novel ligands for mouse olfactory receptor MOR42-3 using an in silico screening approach and in vitro validation. PLoS ONE 2014, 9, e92064. [Google Scholar] [CrossRef] [PubMed]
- Boyle, S.M.; McInally, S.; Ray, A. Expanding the olfactory code by in silico decoding of odor-receptor chemical space. eLife 2013, 2, e01120. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Dahoun, T.; Brugarolas, M.; Pick, H.; Filipek, S.; Vogel, H. Computational modeling of the olfactory receptor Olfr73 suggests a molecular basis for low potency of olfactory receptor-activating compounds. Commun. Biol. 2019, 2, 141. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.; Ren, W.; Pacalon, J.; Xu, R.; Xu, L.; Li, X.; de March, C.A.; Matsunami, H.; Yu, H.; Yu, Y.; et al. Large-Scale G Protein-Coupled Olfactory Receptor-Ligand Pairing. ACS Cent. Sci. 2022, 8, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Mittal, A.; Agrawal, V.; Gupta, S.; Gupta, K.; Jain, R.R.; Garg, P.; Mohanty, S.K.; Sogani, R.; Chhabra, H.S.; et al. OdoriFy: A conglomerate of artificial intelligence-driven prediction engines for olfactory decoding. J. Biol. Chem. 2021, 297, 100956. [Google Scholar] [CrossRef] [PubMed]
- Lalis, M.; Hladis, M.; Khalil, S.A.; Briand, L.; Fiorucci, S.; Topin, J. M2OR: A database of olfactory receptor-odorant pairs for understanding the molecular mechanisms of olfaction. Nucleic Acids Res. 2024, 52, D1370–D1379. [Google Scholar] [CrossRef] [PubMed]
- Kornbausch, N.; Debong, M.W.; Buettner, A.; Heydel, J.M.; Loos, H.M. Odorant Metabolism in Humans. Angew. Chem. Int. Ed. Engl. 2022, 61, e202202866. [Google Scholar] [CrossRef] [PubMed]
- Raka, R.N.; Wu, H.; Xiao, J.; Hossen, I.; Cao, Y.; Huang, M.; Jin, J. Human ectopic olfactory receptors and their food originated ligands: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 5424–5443. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Wang, Y.; Kang, S.G.; Huang, K. Ectopic Odorant Receptor Responding to Flavor Compounds: Versatile Roles in Health and Disease. Pharmaceutics 2021, 13, 1314. [Google Scholar] [CrossRef] [PubMed]
- Bienenstock, J.; Kunze, W.A.; Forsythe, P. Disruptive physiology: Olfaction and the microbiome-gut-brain axis. Biol. Rev. Camb. Philos. Soc. 2018, 93, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Pluznick, J.L.; Protzko, R.J.; Gevorgyan, H.; Peterlin, Z.; Sipos, A.; Han, J.; Brunet, I.; Wan, L.X.; Rey, F.; Wang, T.; et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 4410–4415. [Google Scholar] [CrossRef] [PubMed]
- Pace, U.; Hanski, E.; Salomon, Y.; Lancet, D. Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature 1985, 316, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Sklar, P.B.; Anholt, R.R.; Snyder, S.H. The odorant-sensitive adenylate cyclase of olfactory receptor cells. Differential stimulation by distinct classes of odorants. J. Biol. Chem. 1986, 261, 15538–15543. [Google Scholar] [CrossRef] [PubMed]
- Belluscio, L.; Gold, G.H.; Nemes, A.; Axel, R. Mice deficient in G(olf) are anosmic. Neuron 1998, 20, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Brunet, L.J.; Gold, G.H.; Ngai, J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron 1996, 17, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; LeBel, R.P.; Storm, D.R.; Chen, X. Type 3 adenylyl cyclase: A key enzyme mediating the cAMP signaling in neuronal cilia. Int. J. Physiol. Pathophysiol. Pharmacol. 2016, 8, 95–108. [Google Scholar] [PubMed]
- Meyer, M.R.; Angele, A.; Kremmer, E.; Kaupp, U.B.; Muller, F. A cGMP-signaling pathway in a subset of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 2000, 97, 10595–10600. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lane, A.P.; Bock, R.; Leinders-Zufall, T.; Zufall, F. Blocking adenylyl cyclase inhibits olfactory generator currents induced by “IP(3)-odors”. J. Neurophysiol. 2000, 84, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Ferrand, N.; Pessah, M.; Frayon, S.; Marais, J.; Garel, J.M. Olfactory receptors, Golf alpha and adenylyl cyclase mRNA expressions in the rat heart during ontogenic development. J. Mol. Cell. Cardiol. 1999, 31, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Aisenberg, W.H.; Huang, J.; Zhu, W.; Rajkumar, P.; Cruz, R.; Santhanam, L.; Natarajan, N.; Yong, H.M.; De Santiago, B.; Oh, J.J.; et al. Defining an olfactory receptor function in airway smooth muscle cells. Sci. Rep. 2016, 6, 38231. [Google Scholar] [CrossRef] [PubMed]
- Pluznick, J.L.; Zou, D.J.; Zhang, X.; Yan, Q.; Rodriguez-Gil, D.J.; Eisner, C.; Wells, E.; Greer, C.A.; Wang, T.; Firestein, S.; et al. Functional expression of the olfactory signaling system in the kidney. Proc. Natl. Acad. Sci. USA 2009, 106, 2059–2064. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019, 18, 116–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, L.; Li, H. Role of ectopic olfactory receptors in glucose and lipid metabolism. Br. J. Pharmacol. 2021, 178, 4792–4807. [Google Scholar] [CrossRef] [PubMed]
- Hoover, K.C. Evolution of olfactory receptors. Methods Mol. Biol. 2013, 1003, 241–249. [Google Scholar] [PubMed]
- Niimura, Y.; Nei, M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. USA 2005, 102, 6039–6044. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.B. The molecular architecture of odor and pheromone sensing in mammals. Cell 2000, 100, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Young, J.M.; Trask, B.J. The sense of smell: Genomics of vertebrate odorant receptors. Hum. Mol. Genet. 2002, 11, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Firestein, S. Comparative genomics of odorant and pheromone receptor genes in rodents. Genomics 2007, 89, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Glusman, G.; Bahar, A.; Sharon, D.; Pilpel, Y.; White, J.; Lancet, D. The olfactory receptor gene superfamily: Data mining, classification, and nomenclature. Mamm. Genome 2000, 11, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Glusman, G.; Yanai, I.; Rubin, I.; Lancet, D. The complete human olfactory subgenome. Genome Res. 2001, 11, 685–702. [Google Scholar] [CrossRef] [PubMed]
- Olender, T.; Jones, T.E.M.; Bruford, E.; Lancet, D. A unified nomenclature for vertebrate olfactory receptors. BMC Evol. Biol. 2020, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Olender, T.; Nativ, N.; Lancet, D. HORDE: Comprehensive resource for olfactory receptor genomics. Methods Mol. Biol. 2013, 1003, 23–38. [Google Scholar] [PubMed]
- Zhang, X.; Firestein, S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002, 5, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Quignon, P.; Giraud, M.; Rimbault, M.; Lavigne, P.; Tacher, S.; Morin, E.; Retout, E.; Valin, A.S.; Lindblad-Toh, K.; Nicolas, J.; et al. The dog and rat olfactory receptor repertoires. Genome Biol. 2005, 6, R83. [Google Scholar] [CrossRef] [PubMed]
- Feldmesser, E.; Olender, T.; Khen, M.; Yanai, I.; Ophir, R.; Lancet, D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics 2006, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; De la Cruz, O.; Pinto, J.M.; Nicolae, D.; Firestein, S.; Gilad, Y. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007, 8, R86. [Google Scholar] [CrossRef] [PubMed]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef] [PubMed]
- Jovancevic, N.; Dendorfer, A.; Matzkies, M.; Kovarova, M.; Heckmann, J.C.; Osterloh, M.; Boehm, M.; Weber, L.; Nguemo, F.; Semmler, J.; et al. Medium-chain fatty acids modulate myocardial function via a cardiac odorant receptor. Basic Res. Cardiol. 2017, 112, 13. [Google Scholar] [CrossRef] [PubMed]
- Regard, J.B.; Sato, I.T.; Coughlin, S.R. Anatomical profiling of G protein-coupled receptor expression. Cell 2008, 135, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Phan, J.H.; Wang, M.D. Effect of low-expression gene filtering on detection of differentially expressed genes in RNA-seq data. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 6461–6464. [Google Scholar]
- Ashraf, S.; Frazier, O.H.; Carranza, S.; McPherson, D.D.; Taegtmeyer, H.; Harmancey, R. A Two-Step Transcriptome Analysis of the Human Heart Reveals Broad and Disease-Responsive Expression of Ectopic Olfactory Receptors. Int. J. Mol. Sci. 2023, 24, 13709. [Google Scholar] [CrossRef] [PubMed]
- Kalbe, B.; Knobloch, J.; Schulz, V.M.; Wecker, C.; Schlimm, M.; Scholz, P.; Jansen, F.; Stoelben, E.; Philippou, S.; Hecker, E.; et al. Olfactory Receptors Modulate Physiological Processes in Human Airway Smooth Muscle Cells. Front. Physiol. 2016, 7, 339. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yoon, Y.C.; Lee, A.S.; Kang, N.; Koo, J.; Rhyu, M.R.; Park, J.H. Expression of human olfactory receptor 10J5 in heart aorta, coronary artery, and endothelial cells and its functional role in angiogenesis. Biochem. Biophys. Res. Commun. 2015, 460, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Matsunami, H.; Ley, K. Olfactory receptors in macrophages and inflammation. Front. Immunol. 2022, 13, 1029244. [Google Scholar] [CrossRef] [PubMed]
- Son, B.; Kang, W.; Park, S.; Choi, D.; Park, T. Dermal Olfactory Receptor OR51B5 Is Essential for Survival and Collagen Synthesis in Human Dermal Fibroblast (Hs68 Cells). Int. J. Mol. Sci. 2021, 22, 9273. [Google Scholar] [CrossRef] [PubMed]
- Grosmaitre, X.; Vassalli, A.; Mombaerts, P.; Shepherd, G.M.; Ma, M. Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: A patch clamp analysis in gene-targeted mice. Proc. Natl. Acad. Sci. USA 2006, 103, 1970–1975. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Ryu, S.E.; Min, Y.; de March, C.A.; Bushdid, C.; Golebiowski, J.; Moon, C.; Park, T. Olfactory receptor 10J5 responding to alpha-cedrene regulates hepatic steatosis via the cAMP-PKA pathway. Sci. Rep. 2017, 7, 9471. [Google Scholar] [CrossRef] [PubMed]
- Pluznick, J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes 2014, 5, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Billesbolle, C.B.; de March, C.A.; van der Velden, W.J.C.; Ma, N.; Tewari, J.; Del Torrent, C.L.; Li, L.; Faust, B.; Vaidehi, N.; Matsunami, H.; et al. Structural basis of odorant recognition by a human odorant receptor. Nature 2023, 615, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Mermer, P.; Strotmann, J.; Kummer, W.; Paddenberg, R. Olfactory receptor Olfr78 (prostate-specific G protein-coupled receptor PSGR) expression in arterioles supplying skeletal and cardiac muscles and in arterioles feeding some murine organs. Histochem. Cell Biol. 2021, 156, 539–553. [Google Scholar] [CrossRef]
- Poll, B.G.; Xu, J.; Gupta, K.; Shubitowski, T.B.; Pluznick, J.L. Olfactory receptor 78 modulates renin but not baseline blood pressure. Physiol. Rep. 2021, 9, e15017. [Google Scholar] [CrossRef]
- Morrell, C.N.; Mix, D.; Aggarwal, A.; Bhandari, R.; Godwin, M.; Owens, P., 3rd; Lyden, S.P.; Doyle, A.; Krauel, K.; Rondina, M.T.; et al. Platelet olfactory receptor activation limits platelet reactivity and growth of aortic aneurysms. J. Clin. Investig. 2022, 132, e152373. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Oliveira, V.; Foresto-Neto, O.; Watanabe, I.K.M.; Zatz, R.; Camara, N.O.S. Inflammation in Renal Diseases: New and Old Players. Front. Pharmacol. 2019, 10, 1192. [Google Scholar] [CrossRef] [PubMed]
- Ruparelia, N.; Chai, J.T.; Fisher, E.A.; Choudhury, R.P. Inflammatory processes in cardiovascular disease: A route to targeted therapies. Nat. Rev. Cardiol. 2017, 14, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Chavakis, T.; Cosentino, F.; Schmidt, A.M. Cardiometabolic disease: Linking pathogenic mechanisms to therapeutic opportunities. Cardiovasc. Res. 2024, 119, 2771–2773. [Google Scholar] [CrossRef] [PubMed]
- Guardia, G.D.A.; Naressi, R.G.; Buzzato, V.C.; da Costa, J.B.; Zalcberg, I.; Ramires, J.; Malnic, B.; Gutiyama, L.M.; Galante, P.A.F. Acute Myeloid Leukemia Expresses a Specific Group of Olfactory Receptors. Cancers 2023, 15, 3073. [Google Scholar] [CrossRef] [PubMed]
- Manteniotis, S.; Wojcik, S.; Brauhoff, P.; Mollmann, M.; Petersen, L.; Gothert, J.R.; Schmiegel, W.; Duhrsen, U.; Gisselmann, G.; Hatt, H. Functional characterization of the ectopically expressed olfactory receptor 2AT4 in human myelogenous leukemia. Cell Death Discov. 2016, 2, 15070. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Tay, H.L.; Plank, M.; Essilfie, A.T.; Hansbro, P.M.; Foster, P.S.; Yang, M. Activation of olfactory receptors on mouse pulmonary macrophages promotes monocyte chemotactic protein-1 production. PLoS ONE 2013, 8, e80148. [Google Scholar] [CrossRef] [PubMed]
- Vadevoo, S.M.P.; Gunassekaran, G.R.; Lee, C.; Lee, N.; Lee, J.; Chae, S.; Park, J.Y.; Koo, J.; Lee, B. The macrophage odorant receptor Olfr78 mediates the lactate-induced M2 phenotype of tumor-associated macrophages. Proc. Natl. Acad. Sci. USA 2021, 118, e210243411. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.J.; Ortega, F.E.; Riegler, J.; Madison, D.V.; Krasnow, M.A. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 2015, 527, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Geng, X.; Hou, J.; Wu, G. New insights into M1/M2 macrophages: Key modulators in cancer progression. Cancer Cell Int. 2021, 21, 389. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Peng, W.B.; Zhang, P.; Yang, X.P.; Zhou, Q. Lactate in the tumour microenvironment: From immune modulation to therapy. EBioMedicine 2021, 73, 103627. [Google Scholar] [CrossRef]
- Leblond, A.L.; Klinkert, K.; Martin, K.; Turner, E.C.; Kumar, A.H.; Browne, T.; Caplice, N.M. Systemic and Cardiac Depletion of M2 Macrophage through CSF-1R Signaling Inhibition Alters Cardiac Function Post Myocardial Infarction. PLoS ONE 2015, 10, e0137515. [Google Scholar] [CrossRef] [PubMed]
- Mouton, A.J.; DeLeon-Pennell, K.Y.; Rivera Gonzalez, O.J.; Flynn, E.R.; Freeman, T.C.; Saucerman, J.J.; Garrett, M.R.; Ma, Y.; Harmancey, R.; Lindsey, M.L. Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res. Cardiol. 2018, 113, 26. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, K.C.; Zhang, Y.; Sturgeon, K.; Sinoway, L.I.; Trifiletti, D.M.; Chinchilli, V.M.; Zaorsky, N.G. Fatal heart disease among cancer patients. Nat. Commun. 2020, 11, 2011. [Google Scholar] [CrossRef] [PubMed]
- Frydland, M.; Moller, J.E.; Wiberg, S.; Lindholm, M.G.; Hansen, R.; Henriques, J.P.S.; Moller-Helgestad, O.K.; Bang, L.E.; Frikke-Schmidt, R.; Goetze, J.P.; et al. Lactate is a Prognostic Factor in Patients Admitted With Suspected ST-Elevation Myocardial Infarction. Shock 2019, 51, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef] [PubMed]
- Stanley, W.C. Myocardial energy metabolism during ischemia and the mechanisms of metabolic therapies. J. Cardiovasc. Pharmacol. Ther. 2004, 9 (Suppl. S1), S31–S45. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.J.; Sheedy, F.J.; Fisher, E.A. Macrophages in atherosclerosis: A dynamic balance. Nat. Rev. Immunol. 2013, 13, 709–721. [Google Scholar] [CrossRef] [PubMed]
- McArdle, S.; Buscher, K.; Ghosheh, Y.; Pramod, A.B.; Miller, J.; Winkels, H.; Wolf, D.; Ley, K. Migratory and Dancing Macrophage Subsets in Atherosclerotic Lesions. Circ. Res. 2019, 125, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Kobiyama, K.; Winkels, H.; Ghosheh, Y.; McArdle, S.; Mikulski, Z.; Kiosses, W.B.; Fan, Z.; Wen, L.; Jung, Y.; et al. Olfactory receptor 2 in vascular macrophages drives atherosclerosis by NLRP3-dependent IL-1 production. Science 2022, 375, 214–221. [Google Scholar] [CrossRef]
- Poll, B.G.; Chen, L.; Chou, C.L.; Raghuram, V.; Knepper, M.A. Landscape of GPCR expression along the mouse nephron. Am. J. Physiol. Renal Physiol. 2021, 321, F50–F68. [Google Scholar] [CrossRef]
- Shepard, B.D. The Sniffing Kidney: Roles for Renal Olfactory Receptors in Health and Disease. Kidney360 2021, 2, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Peng, Y.J.; Su, X.; Zhang, C.; Nagati, J.S.; Garcia, J.A.; Prabhakar, N.R. Olfactory receptor 78 regulates erythropoietin and cardiorespiratory responses to hypobaric hypoxia. J. Appl. Physiol. (1985) 2021, 130, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Kalbe, B.; Schlimm, M.; Wojcik, S.; Philippou, S.; Massberg, D.; Jansen, F.; Scholz, P.; Luebbert, H.; Ubrig, B.; Osterloh, S.; et al. Olfactory signaling components and olfactory receptors are expressed in tubule cells of the human kidney. Arch. Biochem. Biophys. 2016, 610, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Choi, R.; Gupta, K.; Warren, H.R.; Santhanam, L.; Pluznick, J.L. An evolutionarily conserved olfactory receptor is required for sex differences in blood pressure. Sci. Adv. 2024, 10, eadk1487. [Google Scholar] [CrossRef] [PubMed]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Motahharynia, A.; Moein, S.; Kiyanpour, F.; Moradzadeh, K.; Yaqubi, M.; Gheisari, Y. Olfactory receptors contribute to progression of kidney fibrosis. NPJ Syst. Biol. Appl. 2022, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, N.V.; Datta, S.; El-Kersh, K.; Sadikot, R.T.; Ganti, A.K.; Batra, S.K.; Jain, M. GPCRs and fibroblast heterogeneity in fibroblast-associated diseases. FASEB J. 2023, 37, e23101. [Google Scholar] [CrossRef] [PubMed]
- Rule, A.D.; Bergstralh, E.J.; Melton, L.J., 3rd; Li, X.; Weaver, A.L.; Lieske, J.C. Kidney stones and the risk for chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2009, 4, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wei, H.; Huang, G.; Jin, L. CCL7 and olfactory transduction pathway activation play an important role in the formation of CaOx and CaP kidney stones. Front. Genet. 2023, 14, 1267545. [Google Scholar] [CrossRef] [PubMed]
- Ku, E.; Lee, B.J.; Wei, J.; Weir, M.R. Hypertension in CKD: Core Curriculum 2019. Am. J. Kidney Dis. 2019, 74, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Mondal, N.K.; Walther, C.P. Insights into Myocardial Fibrosis in Advanced Chronic Kidney Disease Using Human Tissue. Kidney360 2023, 4, 1531–1533. [Google Scholar] [CrossRef] [PubMed]
- Panizo, S.; Martinez-Arias, L.; Alonso-Montes, C.; Cannata, P.; Martin-Carro, B.; Fernandez-Martin, J.L.; Naves-Diaz, M.; Carrillo-Lopez, N.; Cannata-Andia, J.B. Fibrosis in Chronic Kidney Disease: Pathogenesis and Consequences. Int. J. Mol. Sci. 2021, 22, 408. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Chen, K.; Quan, D.; Kang, L.; Yang, D.; Wu, H.; Yan, M.; Wu, S.; Lv, L.; Zhang, G. The Combination of Scutellaria baicalensis Georgi and Sophora japonica L. ameliorate Renal Function by Regulating Gut Microbiota in Spontaneously Hypertensive Rats. Front. Pharmacol. 2020, 11, 575294. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.N.; Hou, C.Y.; Chang, C.I.; Tain, Y.L. Resveratrol Butyrate Ester Protects Adenine-Treated Rats against Hypertension and Kidney Disease by Regulating the Gut-Kidney Axis. Antioxidants 2021, 11, 83. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, P.; Aisenberg, W.H.; Acres, O.W.; Protzko, R.J.; Pluznick, J.L. Identification and characterization of novel renal sensory receptors. PLoS ONE 2014, 9, e111053. [Google Scholar] [CrossRef] [PubMed]
- Shepard, B.D.; Cheval, L.; Peterlin, Z.; Firestein, S.; Koepsell, H.; Doucet, A.; Pluznick, J.L. A Renal Olfactory Receptor Aids in Kidney Glucose Handling. Sci. Rep. 2016, 6, 35215. [Google Scholar] [CrossRef] [PubMed]
- Shepard, B.D.; Koepsell, H.; Pluznick, J.L. Renal olfactory receptor 1393 contributes to the progression of type 2 diabetes in a diet-induced obesity model. Am. J. Physiol. Renal Physiol. 2019, 316, F372–F381. [Google Scholar] [CrossRef] [PubMed]
- Anker, S.D.; Butler, J.; Filippatos, G.; Ferreira, J.P.; Bocchi, E.; Bohm, M.; Brunner-La Rocca, H.P.; Choi, D.J.; Chopra, V.; Chuquiure-Valenzuela, E.; et al. Empagliflozin in Heart Failure with a Preserved Ejection Fraction. N. Engl. J. Med. 2021, 385, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- McMurray, J.J.V.; Solomon, S.D.; Inzucchi, S.E.; Kober, L.; Kosiborod, M.N.; Martinez, F.A.; Ponikowski, P.; Sabatine, M.S.; Anand, I.S.; Belohlavek, J.; et al. Dapagliflozin in Patients with Heart Failure and Reduced Ejection Fraction. N. Engl. J. Med. 2019, 381, 1995–2008. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Bahk, Y.Y.; Lee, N.; Jae, Y.; Cho, Y.H.; Ku, C.R.; Byun, Y.; Lee, E.J.; Kim, M.S.; Koo, J. Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in alpha-cells of mouse pancreatic islets. Biochem. Biophys. Res. Commun. 2015, 460, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Abaffy, T.; Matsunami, H.; Luetje, C.W. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006, 97, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Munakata, Y.; Yamada, T.; Imai, J.; Takahashi, K.; Tsukita, S.; Shirai, Y.; Kodama, S.; Asai, Y.; Sugisawa, T.; Chiba, Y.; et al. Olfactory receptors are expressed in pancreatic beta-cells and promote glucose-stimulated insulin secretion. Sci. Rep. 2018, 8, 1499. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Chi, Q.; Zhuang, H.; Matsunami, H.; Mainland, J.D. Odor coding by a Mammalian receptor repertoire. Sci. Signal 2009, 2, ra9. [Google Scholar] [CrossRef] [PubMed]
- Leem, J.; Shim, H.M.; Cho, H.; Park, J.H. Octanoic acid potentiates glucose-stimulated insulin secretion and expression of glucokinase through the olfactory receptor in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 2018, 503, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.T.; Stevenson, B.E.; Chester, M.W.; Basit, M.; Daniels, M.B.; Turley, S.D.; McGarry, J.D. The insulinotropic potency of fatty acids is influenced profoundly by their chain length and degree of saturation. J. Clin. Investig. 1997, 100, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Dahoun, T.; Grasso, L.; Vogel, H.; Pick, H. Recombinant expression and functional characterization of mouse olfactory receptor mOR256-17 in mammalian cells. Biochemistry 2011, 50, 7228–7235. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Haddad, R.; Chen, S.; Santos, V.; Luetje, C.W. A broadly tuned mouse odorant receptor that detects nitrotoluenes. J. Neurochem. 2012, 121, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Tazir, B.; Khan, M.; Mombaerts, P.; Grosmaitre, X. The extremely broad odorant response profile of mouse olfactory sensory neurons expressing the odorant receptor MOR256-17 includes trace amine-associated receptor ligands. Eur. J. Neurosci. 2016, 43, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Meijerink, J. The Intestinal Fatty Acid-Enteroendocrine Interplay, Emerging Roles for Olfactory Signaling and Serotonin Conjugates. Molecules 2021, 26, 1416. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Voland, P.; Kunz, L.; Prinz, C.; Gratzl, M. Enterochromaffin cells of the human gut: Sensors for spices and odorants. Gastroenterology 2007, 132, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Ripken, D.; van der Wielen, N.; Wortelboer, H.M.; Meijerink, J.; Witkamp, R.F.; Hendriks, H.F. Nutrient-induced glucagon like peptide-1 release is modulated by serotonin. J. Nutr. Biochem. 2016, 32, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Roberts, F.L.; Cataldo, L.R.; Fex, M. Monoamines’ role in islet cell function and type 2 diabetes risk. Trends Mol. Med. 2023, 29, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Lee, I.S.; Kim, K.H.; Park, J.; Kim, Y.; Choi, J.H.; Choi, J.S.; Jang, H.J. Activation of intestinal olfactory receptor stimulates glucagon-like peptide-1 secretion in enteroendocrine cells and attenuates hyperglycemia in type 2 diabetic mice. Sci. Rep. 2017, 7, 13978. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, L.; Zhang, Y.; Block, E.; Buehl, M.; Corr, M.J.; Cormanich, R.A.; Gundala, S.; Matsunami, H.; O’Hagan, D.; Ozbil, M.; et al. Molecular mechanism of activation of human musk receptors OR5AN1 and OR1A1 by (R)-muscone and diverse other musk-smelling compounds. Proc. Natl. Acad. Sci. USA 2018, 115, E3950–E3958. [Google Scholar] [CrossRef] [PubMed]
- Geithe, C.; Noe, F.; Kreissl, J.; Krautwurst, D. The Broadly Tuned Odorant Receptor OR1A1 is Highly Selective for 3-Methyl-2,4-nonanedione, a Key Food Odorant in Aged Wines, Tea, and Other Foods. Chem. Senses 2017, 42, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Schmiedeberg, K.; Shirokova, E.; Weber, H.P.; Schilling, B.; Meyerhof, W.; Krautwurst, D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J. Struct. Biol. 2007, 159, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.E.; Kang, C.W.; Oh, J.H.; Park, S.H.; Ku, C.R.; Cho, Y.H.; Lee, M.K.; Lee, E.J. Olfactory Receptor OR51E1 Mediates GLP-1 Secretion in Human and Rodent Enteroendocrine L Cells. J. Endocr. Soc. 2018, 2, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Priori, D.; Colombo, M.; Clavenzani, P.; Jansman, A.J.; Lalles, J.P.; Trevisi, P.; Bosi, P. The Olfactory Receptor OR51E1 Is Present along the Gastrointestinal Tract of Pigs, Co-Localizes with Enteroendocrine Cells and Is Modulated by Intestinal Microbiota. PLoS ONE 2015, 10, e0129501. [Google Scholar] [CrossRef]
- Wu, C.; Jeong, M.Y.; Kim, J.Y.; Lee, G.; Kim, J.S.; Cheong, Y.E.; Kang, H.; Cho, C.H.; Kim, J.; Park, M.K.; et al. Activation of ectopic olfactory receptor 544 induces GLP-1 secretion and regulates gut inflammation. Gut Microbes 2021, 13, 1987782. [Google Scholar] [CrossRef]
- Wu, C.; Jia, Y.; Lee, J.H.; Kim, Y.; Sekharan, S.; Batista, V.S.; Lee, S.J. Activation of OR1A1 suppresses PPAR-gamma expression by inducing HES-1 in cultured hepatocytes. Int. J. Biochem. Cell Biol. 2015, 64, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Thach, T.T.; Kim, Y.J.; Lee, S.J. Olfactory receptor 43 reduces hepatic lipid accumulation and adiposity in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Romere, C.; Duerrschmid, C.; Bournat, J.; Constable, P.; Jain, M.; Xia, F.; Saha, P.K.; Del Solar, M.; Zhu, B.; York, B.; et al. Asprosin, a Fasting-Induced Glucogenic Protein Hormone. Cell 2016, 165, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Shan, H.; Chen, L.; Long, A.; Zhang, Y.; Liu, Y.; Jia, L.; Wei, F.; Han, J.; Li, T.; et al. OLFR734 Mediates Glucose Metabolism as a Receptor of Asprosin. Cell Metab. 2019, 30, 319–328.e8. [Google Scholar] [CrossRef] [PubMed]
- Thach, T.T.; Wu, C.; Hwang, K.Y.; Lee, S.J. Azelaic Acid Induces Mitochondrial Biogenesis in Skeletal Muscle by Activation of Olfactory Receptor 544. Front. Physiol. 2020, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Hwang, S.H.; Jia, Y.; Choi, J.; Kim, Y.J.; Choi, D.; Pathiraja, D.; Choi, I.G.; Koo, S.H.; Lee, S.J. Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J. Clin. Investig. 2017, 127, 4118–4123. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Park, J.; Moon, C.; Park, T. Regulation of Adipogenesis and Thermogenesis through Mouse Olfactory Receptor 23 Stimulated by alpha-Cedrene in 3T3-L1 Cells. Nutrients 2018, 10, 1781. [Google Scholar] [CrossRef] [PubMed]
- Jahromi, M.M. Haplotype specific alteration of diabetes MHC risk by olfactory receptor gene polymorphism. Autoimmun. Rev. 2012, 12, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Matiashova, L.; Hoogkamer, A.L.; Timper, K. The Role of the Olfactory System in Obesity and Metabolism in Humans: A Systematic Review and Meta-Analysis. Metabolites 2023, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, Z.; Ge, X.Y.; Gao, M.X.; Meng, R.; Xu, X.; Zhang, Y.Q.; Li, R.Z.; Lin, J.Y.; Tian, Z.M.; et al. Autonomous sensing of the insulin peptide by an olfactory G protein-coupled receptor modulates glucose metabolism. Cell Metab. 2022, 34, 240–255.e10. [Google Scholar] [CrossRef] [PubMed]
- Schiazza, A.R.; Considine, E.G.; Betcher, M.; Shepard, B.D. Loss of renal olfactory receptor 1393 leads to improved glucose homeostasis in a type 1 diabetic mouse model. Physiol. Rep. 2021, 9, e15007. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Cho, H.J.; Lee, C.; Koo, J. Odorant receptors in cancer. BMB Rep. 2022, 55, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Dibattista, M.; Pifferi, S.; Menini, A.; Reisert, J. Alzheimer’s Disease: What Can We Learn from the Peripheral Olfactory System? Front. Neurosci. 2020, 14, 440. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beito, M.R.; Ashraf, S.; Odogwu, D.; Harmancey, R. Role of Ectopic Olfactory Receptors in the Regulation of the Cardiovascular–Kidney–Metabolic Axis. Life 2024, 14, 548. https://doi.org/10.3390/life14050548
Beito MR, Ashraf S, Odogwu D, Harmancey R. Role of Ectopic Olfactory Receptors in the Regulation of the Cardiovascular–Kidney–Metabolic Axis. Life. 2024; 14(5):548. https://doi.org/10.3390/life14050548
Chicago/Turabian StyleBeito, Mitchell R., Sadia Ashraf, Dorcas Odogwu, and Romain Harmancey. 2024. "Role of Ectopic Olfactory Receptors in the Regulation of the Cardiovascular–Kidney–Metabolic Axis" Life 14, no. 5: 548. https://doi.org/10.3390/life14050548
APA StyleBeito, M. R., Ashraf, S., Odogwu, D., & Harmancey, R. (2024). Role of Ectopic Olfactory Receptors in the Regulation of the Cardiovascular–Kidney–Metabolic Axis. Life, 14(5), 548. https://doi.org/10.3390/life14050548






