Deciphering the Systemic Impact of Herbal Medicines on Allergic Rhinitis: A Network Pharmacological Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of AR-Related Herbal Prescriptions and AR-Specific Herbs
2.2. Disease-Related Targets and Pathway Analysis
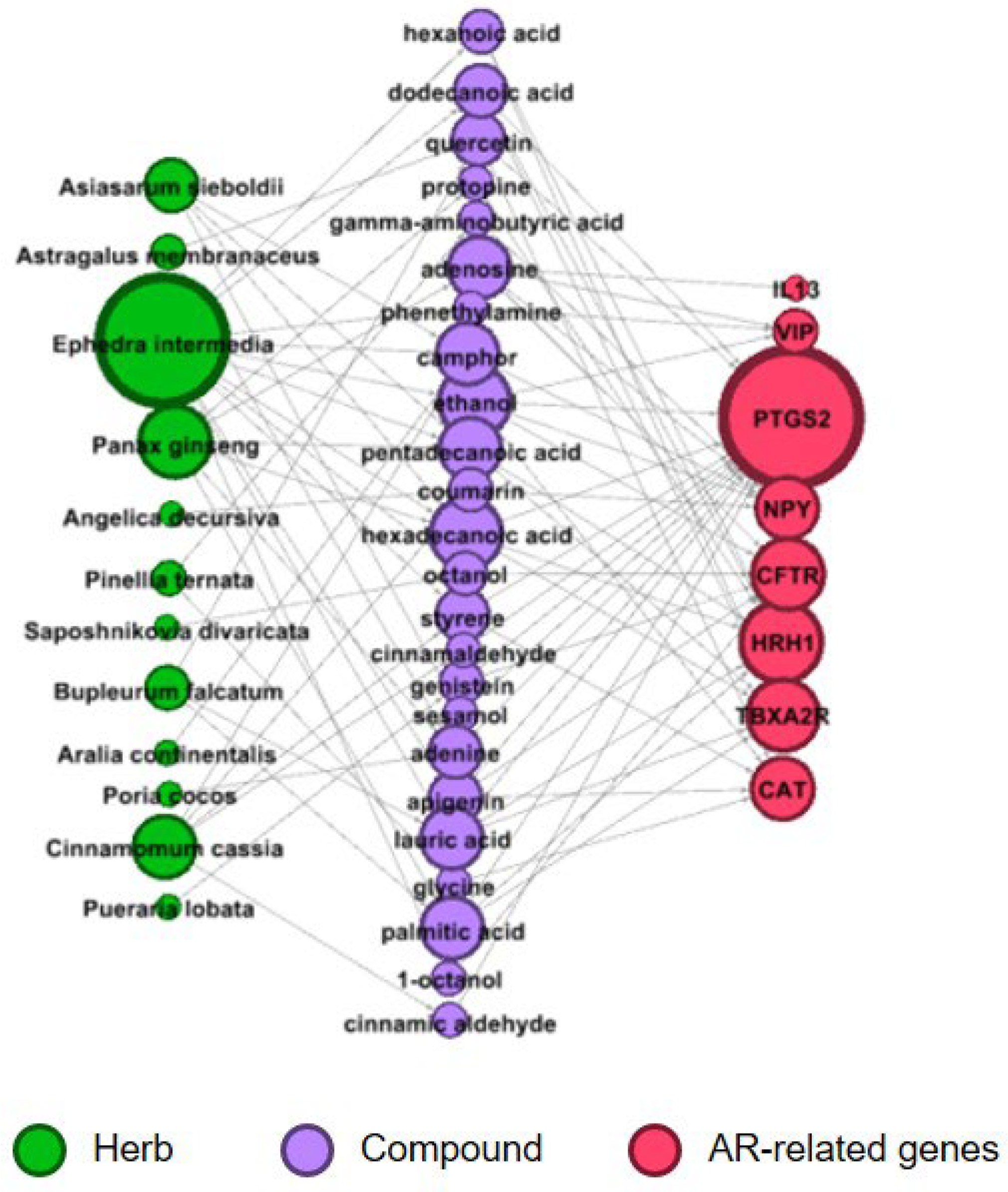
2.3. Construction of an Herb–Compound–Target Network
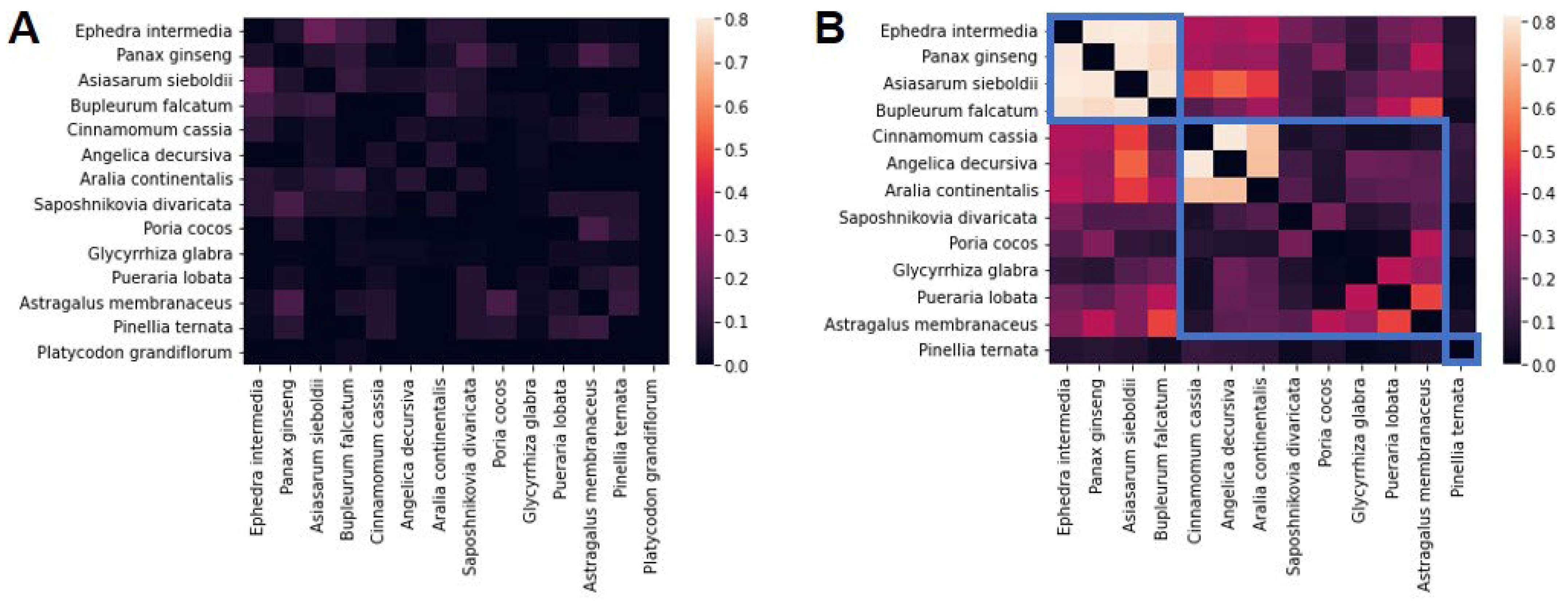
2.4. Identification of Herb Clusters
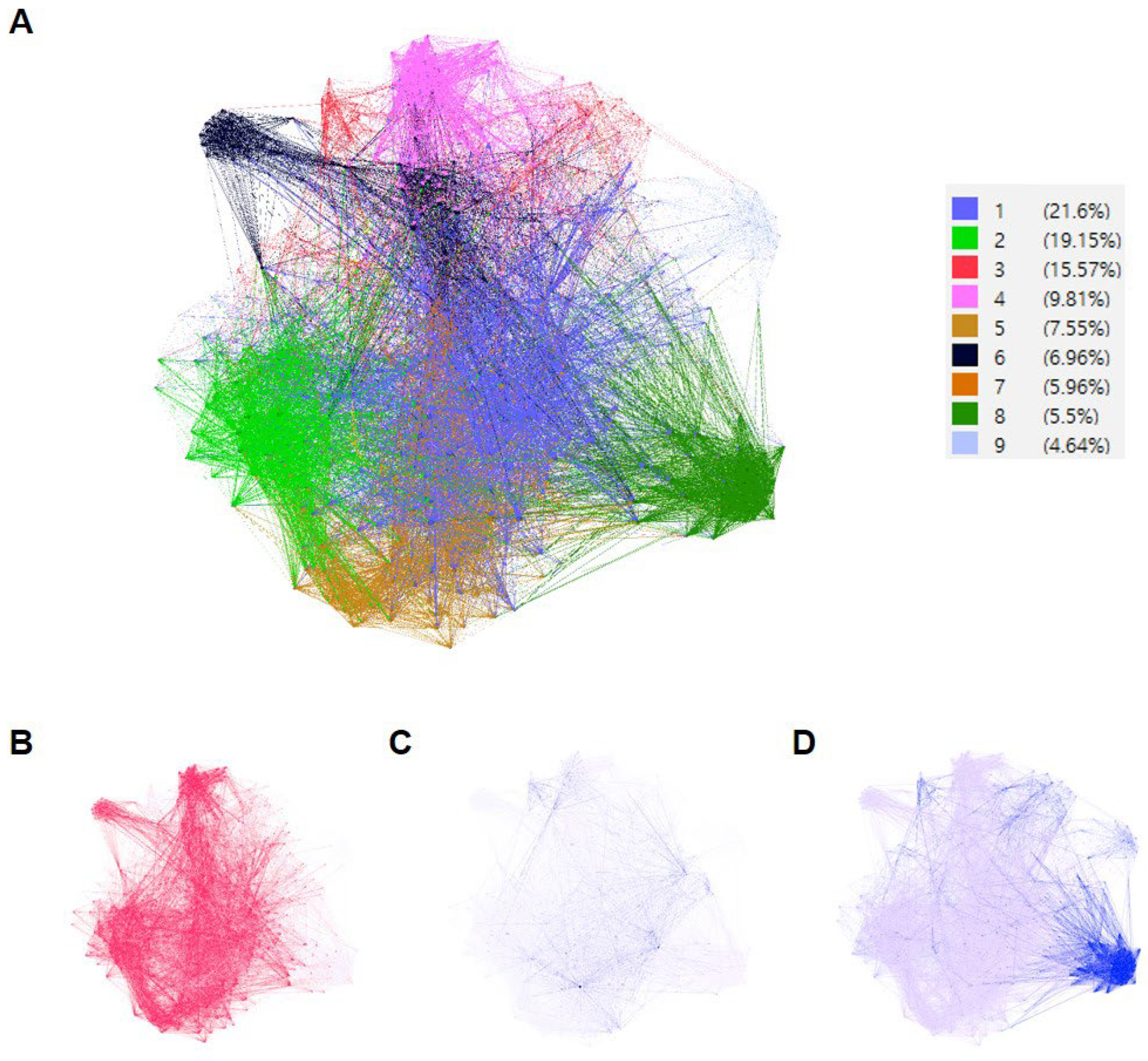
2.5. PPI Network and Gene Ontology (GO) Enrichment Analysis
3. Results
3.1. Identification of AR-Specific Herbs
3.2. AR-Related Genes and Enriched Pathways of AR-Specific Herbs
3.3. Investigation of Groups of AR-Specific Herbs Using PPI Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seidman, M.D.; Gurgel, R.K.; Lin, S.Y.; Schwartz, S.R.; Baroody, F.M.; Bonner, J.R.; Dawson, D.E.; Dykewicz, M.S.; Hackell, J.M.; Han, J.K. Clinical practice guideline: Allergic rhinitis. Otolaryngol.–Head Neck Surg. 2015, 152, S1–S43. [Google Scholar] [CrossRef]
- Ha, J.; Lee, S.W.; Yon, D.K. Ten-Year trends and prevalence of asthma, allergic rhinitis, and atopic dermatitis among the Korean population, 2008–2017. Clin. Exp. Pediatr. 2020, 63, 278. [Google Scholar] [CrossRef]
- Pawankar, R.; Canonica, G.W.; Holgate, S.; Lockey, R. World Allergy Organization White Book on Allergy: Executive Summary 2011–2012; WAO: Milwaukee, WI, USA, 2011. [Google Scholar]
- Yoo, K.-H.; Ahn, H.-R.; Park, J.-K.; Kim, J.-W.; Nam, G.-H.; Hong, S.-K.; Kim, M.-J.; Ghoshal, A.G.; Muttalif, A.R.B.A.; Lin, H.-C. Burden of respiratory disease in Korea: An observational study on allergic rhinitis, asthma, COPD, and rhinosinusitis. Allergy Asthma Immunol. Res. 2016, 8, 527–534. [Google Scholar] [CrossRef]
- Meltzer, E.O.; Bukstein, D.A. The economic impact of allergic rhinitis and current guidelines for treatment. Ann. Allergy Asthma Immunol. 2011, 106, S12–S16. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yoon, S.-J.; Jo, M.-W.; Kim, E.-J.; Kim, H.-J.; Oh, I.-H. Economic burden of allergic rhinitis in Korea. Am. J. Rhinol. Allergy 2010, 24, e110–e113. [Google Scholar] [CrossRef]
- Cingi, C.; Gevaert, P.; Mösges, R.; Rondon, C.; Hox, V.; Rudenko, M.; Muluk, N.; Scadding, G.; Manole, F.; Hupin, C. Multi-morbidities of allergic rhinitis in adults: European academy of allergy and clinical immunology task force report. Clin. Transl. Allergy 2017, 7, 17. [Google Scholar] [CrossRef]
- Feng, L.; Lin, L.; Wang, S.; Zhao, X.; Dai, Q.; Wang, L.; Yang, Y.; Xu, L.; Liu, Y.; An, L. Clinical practice guidelines for the treatment of allergic rhinitis in children with traditional Chinese medicine. Anat. Rec. 2021, 304, 2592–2604. [Google Scholar] [CrossRef]
- Lee, W.-Y.; Lee, C.-Y.; Kim, Y.-S.; Kim, C.-E. The methodological trends of traditional herbal medicine employing network pharmacology. Biomolecules 2019, 9, 362. [Google Scholar] [CrossRef]
- Kim, H.U.; Ryu, J.Y.; Lee, J.O.; Lee, S.Y. A systems approach to traditional oriental medicine. Nat. Biotechnol. 2015, 33, 264–268. [Google Scholar] [CrossRef]
- Jang, D.; Jeong, H.; Kim, C.-E.; Leem, J. A System-Level Mechanism of Anmyungambi Decoction for Obesity: A Network Pharmacological Approach. Biomolecules 2021, 11, 1881. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, B.; Chen, S.; Lin, M.; Chen, Y.; Jin, S.; Chen, W.; Zhang, Y. Applications of network pharmacology in traditional Chinese medicine research. Evid.-Based Complement. Altern. Med. 2020, 2020, 1646905. [Google Scholar] [CrossRef]
- The Society of Korean Medicine Ophthalmology, Otolaryngology & Dermatology. Allergic Rhinitis Clinical Practice Guidelines for Korean Medicine; National Institute for Korean Medicine Development: Gyeongsan, Republic of Korea, 2021; pp. 1–190.
- Huang, C.-W.; Hwang, I.-H.; Yun, Y.-H.; Jang, B.-H.; Chen, F.-P.; Hwang, S.-J.; Ko, S.-G. Population-based comparison of traditional medicine use in adult patients with allergic rhinitis between South Korea and Taiwan. J. Chin. Med. Assoc. 2018, 81, 708–713. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, D833–D839. [Google Scholar] [CrossRef]
- Thorndike, R.L. Who belongs in the family? Psychometrika 1953, 18, 267–276. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Zhang, R.; Yu, S.; Bai, H.; Ning, K. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci. Rep. 2017, 7, 2821. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef]
- Park, J.; Jang, D.; Phung, H.M.; Choi, T.J.; Kim, C.E.; Lee, S.; Kang, K.S.; Choi, S.H. The potential of pharmacological activities of the multi-compound treatment for GERD: Literature review and a network pharmacology-based analysis. Appl. Biol. Chem. 2021, 64, 48. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Kuhn, M.; Szklarczyk, D.; Franceschini, A.; Von Mering, C.; Jensen, L.J.; Bork, P. STITCH 3: Zooming in on protein–chemical interactions. Nucleic Acids Res. 2012, 40, D876–D880. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. 2021. Available online: https://ojs.aaai.org/index.php/ICWSM (accessed on 22 April 2024).
- Murtagh, F.; Legendre, P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement Ward’s criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Ahmad, S.; Azid, N.A.; Boer, J.C.; Lim, J.; Chen, X.; Plebanski, M.; Mohamud, R. The key role of TNF-TNFR2 interactions in the modulation of allergic inflammation: A review. Front. Immunol. 2018, 9, 2572. [Google Scholar] [CrossRef]
- Berings, M.; Gevaert, P.; De Ruyck, N.; Derycke, L.; Holtappels, G.; Pilette, C.; Bachert, C.; Lambrecht, B.N.; Dullaers, M. Fcε RI expression and IgE binding by dendritic cells and basophils in allergic rhinitis and upon allergen immunotherapy. Clin. Exp. Allergy 2018, 48, 970–980. [Google Scholar] [CrossRef]
- Li, Z.; Yu, S.; Jiang, Y.; Fu, Y. Chemokines and chemokine receptors in allergic rhinitis: From mediators to potential therapeutic targets. Eur. Arch. Oto-Rhino-Laryngol. 2022, 279, 5089–5095. [Google Scholar] [CrossRef]
- Matsune, S. Allergic rhinitis and vascular endothelial growth factor. J. Nippon. Med. Sch. 2012, 79, 170–175. [Google Scholar] [CrossRef][Green Version]
- Nam, J.H.; Kim, W.K. The role of TRP channels in allergic inflammation and its clinical relevance. Curr. Med. Chem. 2020, 27, 1446–1468. [Google Scholar] [CrossRef]
- Samra, S.K.; Rajasekaran, A.; Sandford, A.J.; Ellis, A.K.; Tebbutt, S.J. Cholinergic synapse pathway gene polymorphisms associated with late-phase responses in allergic rhinitis. Front. Allergy 2021, 2, 724328. [Google Scholar] [CrossRef]
- Wheatley, L.M.; Togias, A. Allergic rhinitis. New Engl. J. Med. 2015, 372, 456–463. [Google Scholar] [CrossRef]
- Radman, M.; Golshiri, A.; Shamsizadeh, A.; Zainodini, N.; Bagheri, V.; Arababadi, M.; Kennedy, D. Toll-like receptor 4 plays significant roles during allergic rhinitis. Allergol. Immunopathol. 2015, 43, 416–420. [Google Scholar] [CrossRef]
- Prashanta, S.; Choi, S.-G.; Shin, K.-A.; Lee, C.-Y.; Park, J.-I.; Heo, J.-Y.; Im, K.; Park, S.-G. Histidine Suppresses IgE-Mediated Allergic Responses. J. Korean Soc. Food Sci. Nutr. 2018, 47, 697–702. [Google Scholar]
- Wee, J.H.; Zhang, Y.-L.; Rhee, C.-S.; Kim, D.-Y. Inhibition of allergic response by intranasal selective NF-κB decoy oligodeoxynucleotides in a murine model of allergic rhinitis. Allergy Asthma Immunol. Res. 2017, 9, 61–69. [Google Scholar] [CrossRef]
- Ouyang, Y.; Miyata, M.; Hatsushika, K.; Ohnuma, Y.; Katoh, R.; Ogawa, H.; Okumura, K.; Masuyama, K.; Nakao, A. TGF-β signaling may play a role in the development of goblet cell hyperplasia in a mouse model of allergic rhinitis. Allergol. Int. 2010, 59, 313–319. [Google Scholar] [CrossRef]
- Shiraishi, Y.; Jia, Y.; Domenico, J.; Joetham, A.; Karasuyama, H.; Takeda, K.; Gelfand, E.W. Sequential engagement of FcεRI on mast cells and basophil histamine H4 receptor and FcεRI in allergic rhinitis. J. Immunol. 2013, 190, 539–548. [Google Scholar] [CrossRef]
- Li, Y.; Gao, J.; Kamran, M.; Harmacek, L.; Danhorn, T.; Leach, S.M.; O’Connor, B.P.; Hagman, J.R.; Huang, H. GATA2 regulates mast cell identity and responsiveness to antigenic stimulation by promoting chromatin remodeling at super-enhancers. Nat. Commun. 2021, 12, 494. [Google Scholar] [CrossRef]
- Ahmadiafshar, A.; Ahmadiafshar, S. Efficacy and safety of inhaled and intranasal corticosteroids. Anti-Inflamm. Anti-Allergy Agents Med. Chem. (Former. Curr. Med. Chem.—Anti-Inflamm. Anti-Allergy Agents) 2014, 13, 83–87. [Google Scholar]
- Bernardini, S.; Tiezzi, A.; Laghezza Masci, V.; Ovidi, E. Natural products for human health: An historical overview of the drug discovery approaches. Nat. Prod. Res. 2018, 32, 1926–1950. [Google Scholar] [CrossRef]
- Lim, S.; Jeong, I.; Cho, J.; Shin, C.; Kim, K.-I.; Shim, B.-S.; Ko, S.-G.; Kim, B. The Natural Products Targeting on Allergic Rhinitis: From Traditional Medicine to Modern Drug Discovery. Antioxidants 2021, 10, 1524. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, Y.; Li, Y.; Xu, L.; Wang, Y.; Guo, Z.; Song, H.; Yang, M.; Luo, B.; Zheng, A. Ephedrine hydrochloride inhibits PGN-induced inflammatory responses by promoting IL-10 production and decreasing proinflammatory cytokine secretion via the PI3K/Akt/GSK3β pathway. Cell. Mol. Immunol. 2013, 10, 330–337. [Google Scholar] [CrossRef]
- He, W.; Ma, J.; Chen, Y.; Jiang, X.; Wang, Y.; Shi, T.; Zhang, Q.; Yang, Y.; Jiang, X.; Yin, S. Ephedrine hydrochloride protects mice from staphylococcus aureus-induced peritonitis. Am. J. Transl. Res. 2018, 10, 670. [Google Scholar]
- Wang, D.; Bu, T.; Li, Y.; He, Y.; Yang, F.; Zou, L. Pharmacological activity, pharmacokinetics, and clinical research progress of puerarin. Antioxidants 2022, 11, 2121. [Google Scholar] [CrossRef]
- Tong, J.; Hu, X.J.; Cai, W.Q.; Dai, X.; Wang, L. Puerarin alleviates delayed-type hypersensitivity via cytokine inhibition by modulating Th1/Th2 balance. Exp. Ther. Med. 2018, 15, 4441–4447. [Google Scholar] [CrossRef]
- Du, W.; Su, J.; Ye, D.; Wang, Y.; Huang, Q.; Gong, X. Pinellia ternata attenuates mucus secretion and airway inflammation after inhaled corticosteroid withdrawal in COPD rats. Am. J. Chin. Med. 2016, 44, 1027–1041. [Google Scholar] [CrossRef]
- Zou, T.; Wang, J.; Wu, X.; Yang, K.; Zhang, Q.; Wang, C.; Wang, X.; Zhao, C. A review of the research progress on Pinellia ternata (Thunb.) Breit.: Botany, traditional uses, phytochemistry, pharmacology, toxicity and quality control. Heliyon 2023, 9, e22153. [Google Scholar] [CrossRef]
- Martín-Vázquez, E.; Cobo-Vuilleumier, N.; López-Noriega, L.; Lorenzo, P.I.; Gauthier, B.R. The PTGS2/COX2-PGE2 signaling cascade in inflammation: Pro or anti? A case study with type 1 diabetes mellitus. Int. J. Biol. Sci. 2023, 19, 4157. [Google Scholar] [CrossRef]
- Chu, J.T. Histamine H1 receptor gene polymorphism acts as a biological indicator of the prediction of therapeutic efficacy in patients with allergic rhinitis in the Chinese Han population. J. Cell. Biochem. 2019, 120, 164–170. [Google Scholar] [CrossRef]
- Rucker, D.; Dhamoon, A.S. Physiology, Thromboxane A2; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Yong, M.; Hernaiz-Leonardo, J.C.; Alqunaee, M.; Quon, B.S.; Javer, A. The prevalence of CFTR mutations in patients with chronic rhinosinusitis: A systematic review and meta-analysis. Clin. Otolaryngol. 2022, 47, 24–33. [Google Scholar] [CrossRef]
- Rydzewski, B.; Pruszewicz, A.; Sulkowski, W.J. Assessment of smell and taste in patients with allergic rhinitis. Acta Oto-Laryngol. 2000, 120, 323–326. [Google Scholar]
- Kook, J.; Kim, H.; Kim, H.; Kim, K.; Kim, T.; Kang, K.; Oh, D.; Lee, S.H. Increased expression of bitter taste receptors in human allergic nasal mucosa and their contribution to the shrinkage of human nasal mucosa. Clin. Exp. Allergy 2016, 46, 584–601. [Google Scholar] [CrossRef]

| Scientific Name | Latin Name | Chinese Name |
|---|---|---|
| Ephedra intermedia | Ephedra Herba | Ma Huang (麻黃) |
| Pueraria lobata | Puerariae Radix | Ge Gen (葛根) |
| Poria cocos | Poria Sclerotium | Fu Ling (茯苓) |
| Pinellia ternata | Pinelliae Tuber | Ban Xia (半夏) |
| Glycyrrhiza glabra | Glycyrrhizae Radix et Rhizoma | Gan Cao (甘草) |
| Asiasarum sieboldii | Asiasari Radix et Rhizoma | Xi Xin (細辛) |
| Cinnamomum cassia | Cinnamomi Ramulus | Gui Zhi (肉桂) |
| Bupleurum falcatum | Bupleuri Radix | Chai Hu (柴胡) |
| Platycodon grandiflorum | Platycodonis Radix | Jie Geng (桔梗) |
| Astragalus membranaceus | Astragali Radix | Huang Qi (黃芪) |
| Angelica decursiva | Peucedani Radix | Qian Hu (前胡) |
| Angelica pubescens | Angelica Pubescens Radix | Du Huo (獨活) |
| Panax ginseng | Ginseng Radix | Ren Shen (人蔘) |
| Saposhnikovia divaricata | Saposhnikoviae Radix | Fang Feng (防風) |
| Herb name | Number of H-C Interactions | Number of C-T Interactions | Number of Overlapped Genes (Interactions) Between Targets of AR-Specific Herbs and AR-Related Genes | Overlapped Gene Name (Number of C-T Interactions) |
|---|---|---|---|---|
| Ephedra intermedia | 106 | 2021 | 7(27) | CAT(3), PTGS2(6), HRH1(5), TBXA2R(5), VIP(2), NPY(2), CFTR(4) |
| Pueraria lobata | 16 | 207 | 2 | PTGS2, CFTR |
| Poria cocos | 13 | 122 | 1 | PTGS2 |
| Pinellia ternata | 25 | 1553 | 2 | CAT, NPY |
| Glycyrrhiza glabra | 82 | 74 | 0 | - |
| Asiasarum sieboldii | 73 | 914 | 4(7) | PTGS2(2), HRH1(2), TBXA2R(2), NPY |
| Cinnamomum cassia | 18 | 448 | 3(5) | CAT, PTGS2(2), NPY(2) |
| Bupleurum falcatum | 56 | 948 | 4(8) | CAT, PTGS2(3), HRH1(2), TBXA2R(2) |
| Platycodon grandiflorum | 6 | 0 | 0 | - |
| Astragalus membranaceus | 20 | 405 | 2(3) | CAT, PTGS2(2) |
| Angelica decursiva | 16 | 263 | 1 | NPY |
| Aralia continentalis | 79 | 311 | 1 | NPY |
| Panax ginseng | 88 | 1707 | 7(14) | PTGS2(3), IL13, HRH1(4), TBXA2R(3), VIP, NPY, CFTR |
| Saposhnikovia divaricata | 41 | 81 | 1 | CFTR |
| Pathway | Overlap | Adjusted p-Value | Combined Score | ||
|---|---|---|---|---|---|
| KEGG pathway database | |||||
| Cytokine–cytokine receptor interaction | 78/295 | 6.42 × 10−16 | 121.53 | ||
| Previous research | |||||
| Chemokine signaling pathway [29] | 110/192 | 1.47 × 10−58 | 1771.89 | ||
| Cholinergic synapse [32] | 77/113 | 3.55 × 10−49 | 2331.14 | ||
| Inflammatory mediator regulation of TRP channels [31] | 57/98 | 2.95 × 10−31 | 947.24 | ||
| TNF signaling pathway [27] | 49/112 | 3.80 × 10−20 | 334.19 | ||
| VEGF signaling pathway [30] | 34/59 | 4.49 × 10−19 | 548.67 | ||
| IL-17 signaling pathway [33] | 43/94 | 9.56 × 10−19 | 335.07 | ||
| T cell receptor signaling pathway [33] | 40/104 | 1.86 × 10−14 | 188.84 | ||
| Toll-like receptor signaling pathway [34] | 38/104 | 5.26 × 10−13 | 155.29 | ||
| Histidine metabolism [35] | 16/22 | 6.86 × 10−12 | 648.97 | ||
| Th17 cell differentiation [33] | 37/107 | 7.01 × 10−12 | 129.43 | ||
| Antigen processing and presentation [33] | 29/78 | 1.90 × 10−10 | 126.10 | ||
| B cell receptor signaling pathway [33] | 29/81 | 5.16 × 10−10 | 113.37 | ||
| Fc epsilon RI signaling pathway [28,38] | 26/68 | 7.89 × 10−10 | 123.12 | ||
| Th1 and Th2 cell differentiation [33] | 30/92 | 3.25 × 10−09 | 89.90 | ||
| NF-kappa B signaling pathway [36] | 32/104 | 4.80 × 10−09 | 80.94 | ||
| TGF-beta signaling pathway [37] | 17/94 | 0.012365 | 9.30 | ||
| GO Term | Overlap | Adjusted p-Value | Combined Score |
|---|---|---|---|
| Group 1 | |||
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway (GO:0007188) | 83/165 | 1.19 × 10−55 | 2472.19 |
| positive regulation of cytosolic calcium ion concentration (GO:0007204) | 71/147 | 6.81 × 10−46 | 1868.84 |
| retinoid metabolic process (GO:0001523) | 56/92 | 2.22 × 10−43 | 2904.11 |
| negative regulation of inflammatory response (GO:0050728) | 81/212 | 2.67 × 10−43 | 1170.88 |
| regulation of inflammatory response to antigenic stimulus (GO:0002861) | 63/137 | 3.44 × 10−39 | 1442.96 |
| regulation of cytosolic calcium ion concentration (GO:0051480) | 65/148 | 5.14 × 10−39 | 1321.08 |
| inflammatory response (GO:0006954) | 79/230 | 1.86 × 10−38 | 877.027 |
| negative regulation of inflammatory response to antigenic stimulus (GO:0002862) | 62/136 | 1.86 × 10−38 | 1386.64 |
| adenylate cyclase-activating G protein-coupled receptor signaling pathway (GO:0007189) | 57/118 | 4.97 × 10−37 | 1484 |
| phospholipase C-activating G protein-coupled receptor signaling pathway (GO:0007200) | 48/81 | 1.30 × 10−36 | 2266.77 |
| Group 2 | |||
| inflammatory response (GO:0006954) | 55/230 | 1.19 × 10−30 | 867.789 |
| adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway (GO:0007193) | 32/60 | 4.54 × 10−30 | 2968.78 |
| detection of chemical stimulus involved in sensory perception of bitter taste (GO:0001580) | 25/40 | 1.17 × 10−25 | 3676.68 |
| sensory perception of bitter taste (GO:0050913) | 25/41 | 2.19 × 10−25 | 3397.9 |
| detection of chemical stimulus involved in sensory perception of taste (GO:0050912) | 25/44 | 2.20 × 10−24 | 2747.01 |
| cytokine-mediated signaling pathway (GO:0019221) | 76/621 | 1.54 × 10−23 | 300.39 |
| cellular response to cytokine stimulus (GO:0071345) | 66/482 | 5.56 × 10−23 | 329.317 |
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway (GO:0007188) | 40/165 | 9.92 × 10−23 | 634.668 |
| positive regulation of cytosolic calcium ion concentration (GO:0007204) | 37/147 | 1.38 × 10−21 | 631.861 |
| estrogen metabolic process (GO:0008210) | 20/30 | 1.46 × 10−21 | 3647.25 |
| Group 3 | |||
| regulation of cellular amino acid metabolic process (GO:0006521) | 37/54 | 2.92 × 10−40 | 7248.12 |
| regulation of cellular amine metabolic process (GO:0033238) | 36/51 | 3.30 × 10−40 | 7906.43 |
| antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent (GO:0002479) | 41/73 | 3.30 × 10−40 | 4225.02 |
| antigen processing and presentation of exogenous peptide antigen via MHC class I (GO:0042590) | 42/78 | 3.30 × 10−40 | 3849.61 |
| pre-replicative complex assembly (GO:0036388) | 39/64 | 3.30 × 10−40 | 5120.1 |
| negative regulation of cell cycle G2/M phase transition (GO:1902750) | 36/57 | 6.80 × 10−38 | 5281.22 |
| regulation of transcription from RNA polymerase II promoter in response to hypoxia (GO:0061418) | 39/75 | 1.40 × 10−36 | 3236.09 |
| anaphase-promoting complex-dependent catabolic process (GO:0031145) | 40/84 | 1.32 × 10−35 | 2646.42 |
| regulation of cellular ketone metabolic process (GO:0010565) | 36/64 | 1.95 × 10−35 | 3697.92 |
| regulation of transcription from RNA polymerase II promoter in response to stress (GO:0043618) | 40/87 | 6.33 × 10−35 | 2425.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-Y.; Lee, Y.Y.; Kim, M.H.; Kim, C.-E. Deciphering the Systemic Impact of Herbal Medicines on Allergic Rhinitis: A Network Pharmacological Approach. Life 2024, 14, 553. https://doi.org/10.3390/life14050553
Park S-Y, Lee YY, Kim MH, Kim C-E. Deciphering the Systemic Impact of Herbal Medicines on Allergic Rhinitis: A Network Pharmacological Approach. Life. 2024; 14(5):553. https://doi.org/10.3390/life14050553
Chicago/Turabian StylePark, Sa-Yoon, Yoon Yeol Lee, Min Hee Kim, and Chang-Eop Kim. 2024. "Deciphering the Systemic Impact of Herbal Medicines on Allergic Rhinitis: A Network Pharmacological Approach" Life 14, no. 5: 553. https://doi.org/10.3390/life14050553
APA StylePark, S.-Y., Lee, Y. Y., Kim, M. H., & Kim, C.-E. (2024). Deciphering the Systemic Impact of Herbal Medicines on Allergic Rhinitis: A Network Pharmacological Approach. Life, 14(5), 553. https://doi.org/10.3390/life14050553








