Understanding the Thermodynamics of Magnesium Binding to RNA Structural Motifs
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
4.1. Significance of Binding Affinity
4.2. Binding Affinity versus Cellular Concentration of Mg2+(aq)
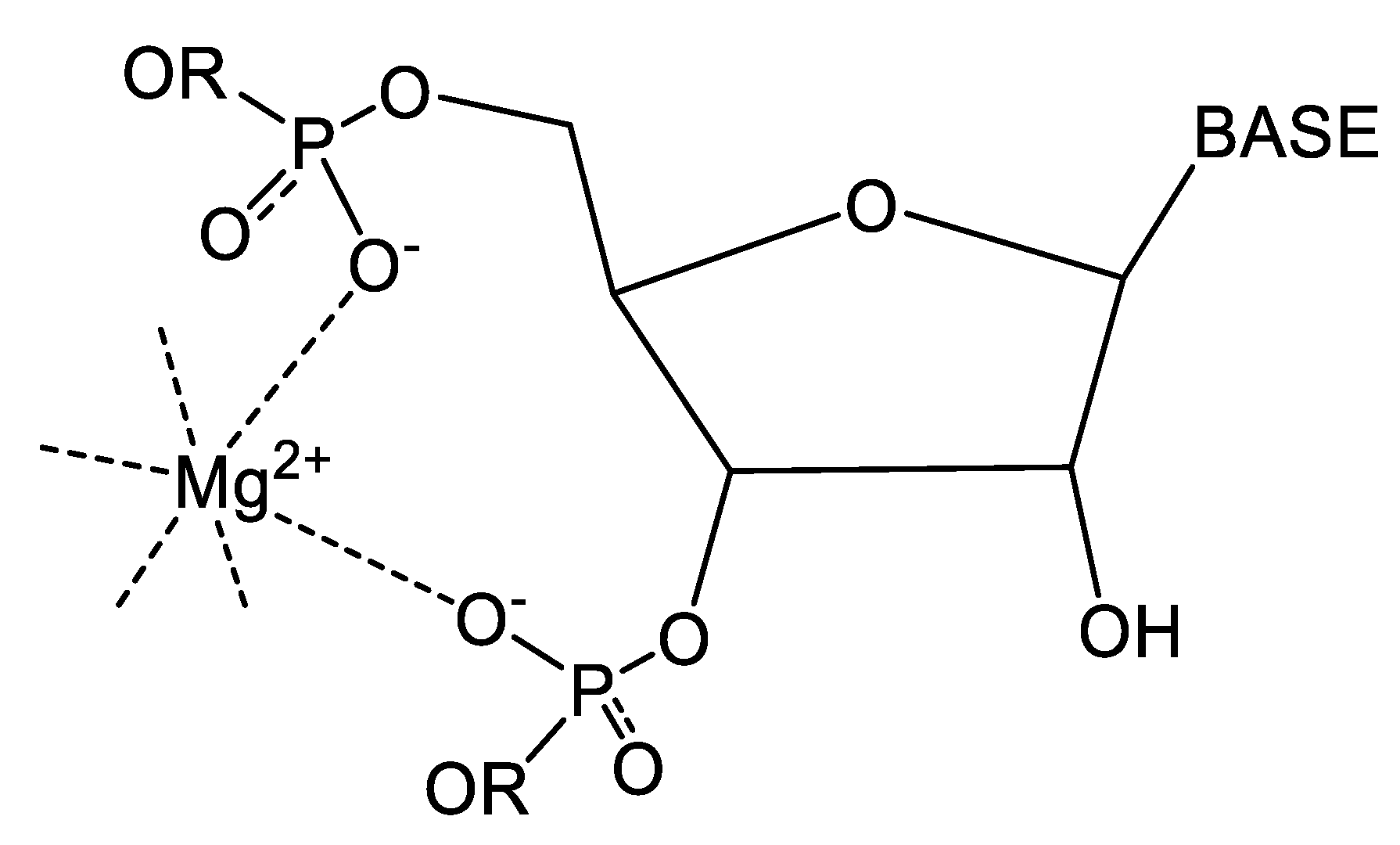
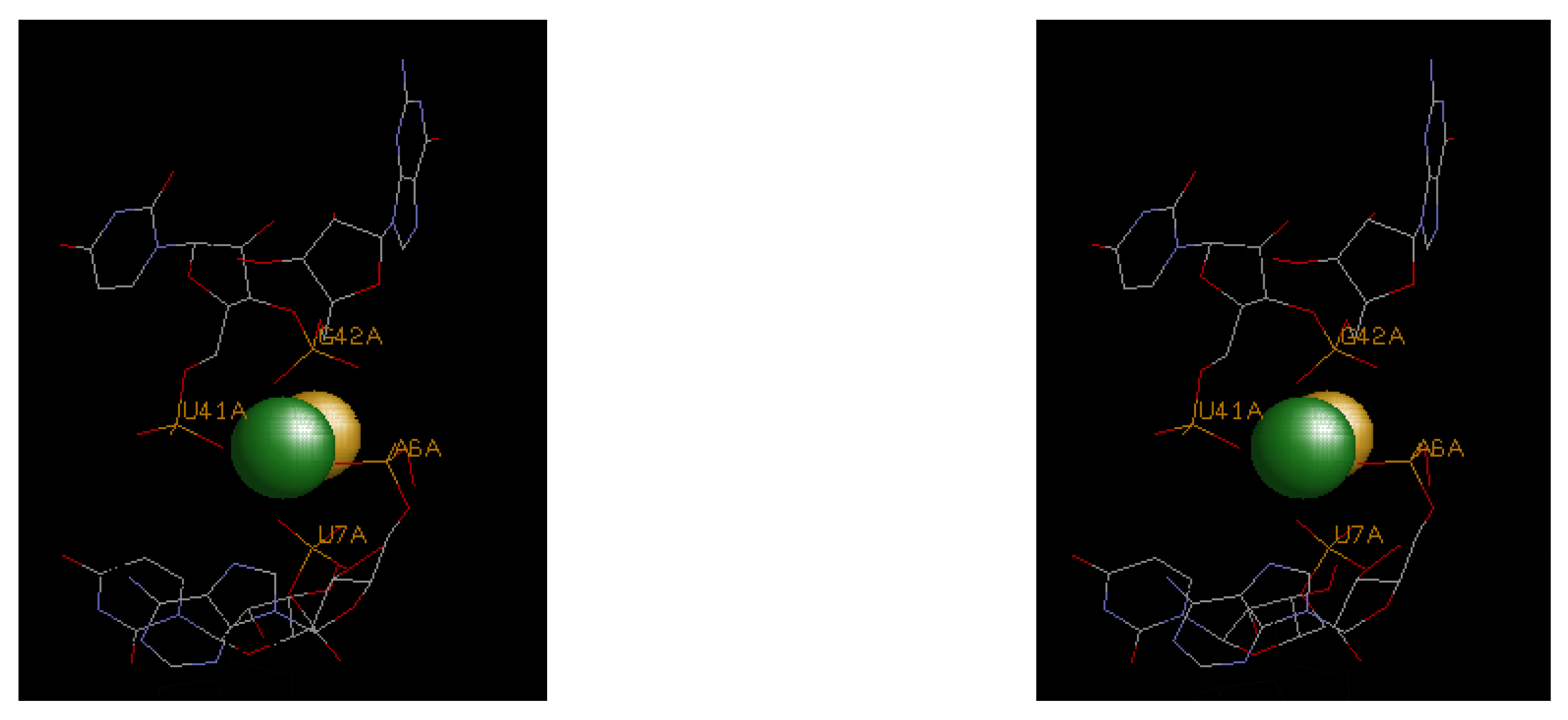
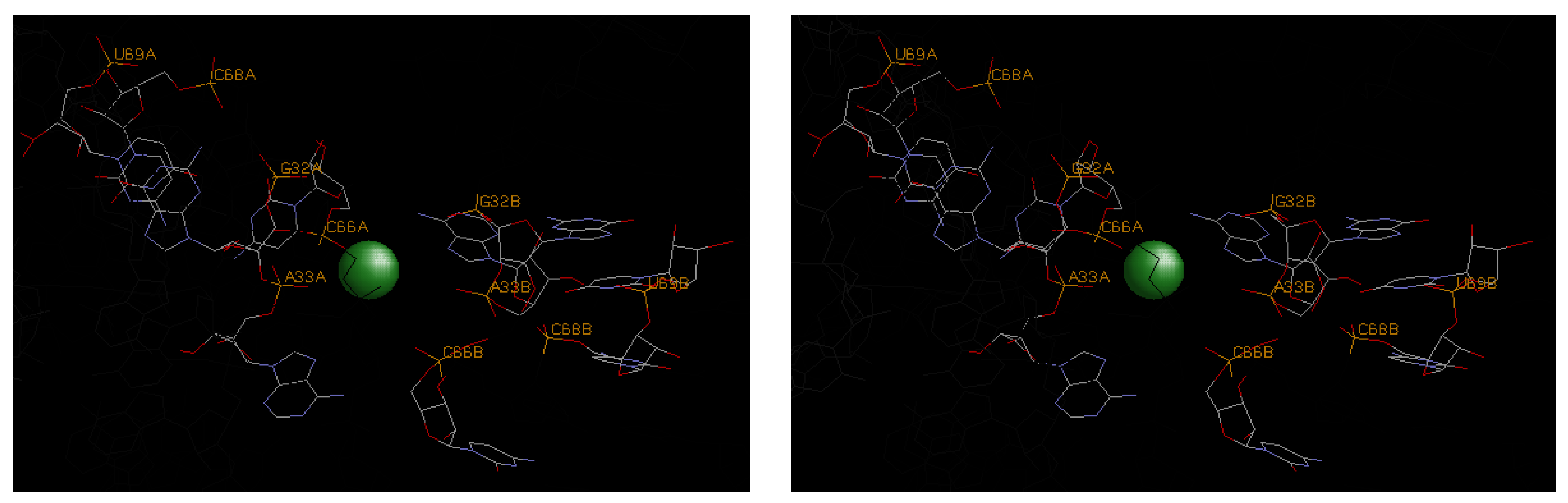
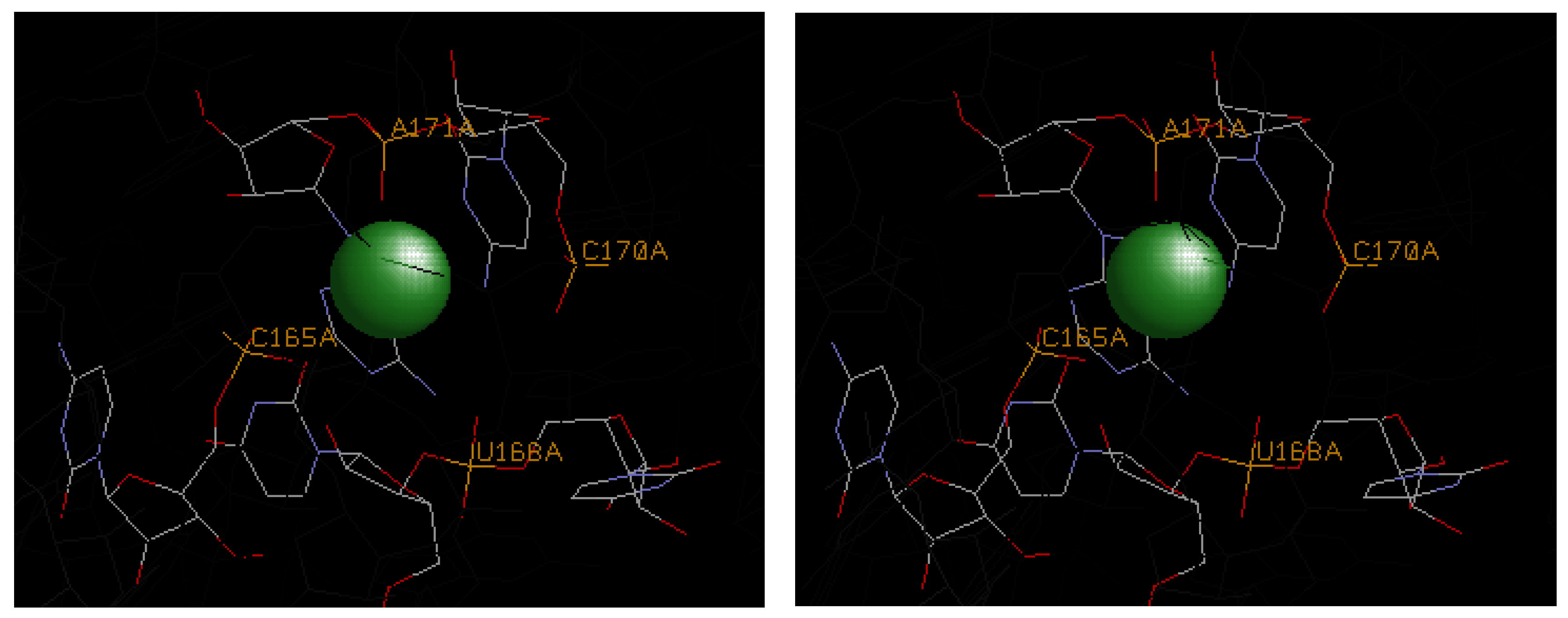
4.3. Illustrative Examples of Magnesium Binding Motifs
4.4. Mg2+ Binding Energetics
4.5. Comparison with Earlier Structural and Thermodynamic Studies of Divalent Magnesium Complexes with RNA
4.6. Caveats
4.7. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Börner, R.; Kowerko, D.; Guiset Miserachs, H.; Schaffer, M.F.; Sigel, R.K.O. Metal ion induced heterogeneity in RNA folding studied by smFRET. Coord. Chem. Rev. 2016, 327–328, 123–142. [Google Scholar] [CrossRef]
- Roy, S.; Lammert, H.; Hayes, R.L.; Chen, B.; LeBlanc, R.; Dayie, T.K.; Onuchic, J.N.; Sanbonmatsu, K.Y. A magnesium-induced triplex pre-organizes the SAM-II riboswitch. PLoS Comput. Biol. 2017, 13, e1005406. [Google Scholar] [CrossRef] [PubMed]
- Yamagami, R.; Bingaman, J.L.; Frankel, E.A.; Bevilacqua, P.C. Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis. Nat. Commun. 2018, 9, 2149. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Chen, S.-J. Hexahydrated Mg2+ Binding and Outer-Shell Dehydration on RNA Surface. Biophys. J. 2018, 114, 1274–1284. [Google Scholar] [CrossRef]
- Zheng, H.; Shabalin, I.G.; Handing, K.B.; Bujnicki, J.M.; Minor, W. Magnesium-binding architectures in RNA crystal structures: Validation, binding preferences, classification and motif detection. Nucleic Acids Res. 2015, 43, 3789–3801. [Google Scholar] [CrossRef]
- Black, C.B.; Huang, H.H.-W.; Cowan, J.A. Biological Coordination Chemistry of Magnesium, Sodium, and Potassium Ions. Protein and Nucleotide Binding Domains. Coord. Chem. Rev. 1994, 135/136, 165–202. [Google Scholar] [CrossRef]
- Cowan, J.A. Coordination chemistry of magnesium ions and 5S rRNA (Escherichia coli): Binding parameters, ligand symmetry, and implications for activity. J. Am. Chem. Soc. 1991, 113, 675–676. [Google Scholar] [CrossRef]
- Cowan, J.A.; Reid, S.S. Biostructural chemistry of magnesium ion: Characterization of the weak binding sites on tRNA(Phe)(yeast). Implications for conformational change and activity. Biochemistry 1990, 29, 6025–6032. [Google Scholar]
- Da Costa, J.B.; Dieckmann, T. Entropy and Mg2+ control ligand affinity and specificity in the malachite green binding RNA aptamer. Mol. BioSyst. 2011, 7, 2156–2163. [Google Scholar] [CrossRef]
- Gonzalez, R.L.J.; Tinoco, I.J. Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J. Mol. Biol. 1999, 289, 1267–1282. [Google Scholar] [CrossRef]
- Kieft, J.S.; Tinoco, I.J. Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine. Structure 1997, 5, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.K.; Draper, D.E. On the role of magnesium ions in RNA stability. Biopolymers 1998, 48, 113–135. [Google Scholar] [CrossRef]
- Misra, V.K.; Draper, D.E. Mg2+ binding to tRNA revisited: The nonlinear poisson-boltzmann model. J. Mol. Biol. 2000, 299, 813–825. [Google Scholar] [CrossRef]
- Misra, V.K.; Draper, D.E. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl. Acad. Sci. USA 2001, 98, 12456–12461. [Google Scholar] [CrossRef]
- Réblová, K.; Spacková, N.; Sponer, J.E.; Koca, J.; Sponer, J. Molecular dynamics simulations of RNA kissing-loop motifs reveal structural dynamics and formation of cation-binding pockets. Nucleic Acids Res. 2003, 31, 6942–6952. [Google Scholar] [CrossRef]
- Reid, S.S.; Cowan, J.A. Metallobiochemistry of a ribosomal RNA. A possible role for Na+ and K+ in the regulation of Mg2+ binding sites on Escherichia coli 5S rRNA: Implications for activity. J. Am. Chem. Soc. 1991, 113, 673–675. [Google Scholar] [CrossRef]
- Saunders, A.M.; DeRose, V.J. Beyond Mg2+: Functional interactions between RNA and transition metals. Curr. Opin. Chem. Biol. 2016, 31, 153–159. [Google Scholar] [CrossRef]
- Black, C.B.; Cowan, J.A. Quantitative Evaluation of Electrostatic and Hydrogen-Bonding Contributions to Metal Cofactor Binding to Nucleic Acids. J. Am. Chem. Soc. 1994, 116, 1174–1178. [Google Scholar] [CrossRef]
- Petrov, A.S.; Bowman, J.C.; Harvey, S.C.; Williams, L.D. Bidentate RNA-Magnesium clamps: On the origin of the special role of magnesium in RNA folding. RNA 2019, 17, 291–297. [Google Scholar] [CrossRef]
- Cowan, J.A. Inorganic Biochemistry. An Introduction, 2nd ed.; Wiley-VCH: New York, NY, USA, 1997. [Google Scholar]
- Bidlack, W.R. The Biological Chemistry of Magnesium; Cowan, J.A., Ed.; VCH: New York, NY, USA, 1995. [Google Scholar]
- Black, C.B.; Cowan, J.A. Inert Chromium and Cobalt Complexes as Probes of Magnesium Dependent Enzymes. Evaluation of the Stoichiometry and Mechanistic Role of the Essential Metal Cofactor in E. coli Exonuclease III. Eur. J. Biochem. 1997, 243, 684–689. [Google Scholar] [CrossRef]
- Dudev, T.; Cowan, J.A.; Lim, C. Competitive Binding in Magnesium Coordination Chemistry: Water versus Ligands of Biological Interest. J. Am. Chem. Soc. 1999, 121, 7665–7673. [Google Scholar] [CrossRef]
- Jou, R.; Cowan, J.A. Ribonuclease H Activation by Inert Transition Metal Complexes. Mechanistic Probes for Metallocofactors: Insights on the Metallobiochemisty of Divalent Magnesium Ion. J. Am. Chem. Soc. 1991, 113, 6685–6686. [Google Scholar] [CrossRef]
- Cunha, R.A.; Bussi, G. Unraveling Mg2+-RNA binding with atomistic dynamics. RNA 2019, 23, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.M.; Poleto, M.D.; Steuer, J.; van der Spoel, D. Influence of Na+ and Mg2+ ions on RNA structures studied with molecular dynamics simulations. Nucleic Acids Res. 2018, 46, 4872–4882. [Google Scholar] [CrossRef]


| ∆Gb (kcal mol−1) Inner Sphere | ∆Gb (kcal mol−1) Outer Sphere |
|---|---|
| −3.3 | −1.2 |
| Motif Name | Type |
|---|---|
| Magnesium clamp | 2 × cis-Pi or 2 × trans-Pi |
| 10-membered ring | 2 or 3 consecutive Pi in cis-, fac-, or mer- geometries (for 3-coordination the third Pi may be separated by one residue from the 10-membered ring), |
| or 4 Pi from two distinct 10-membered-ring pairs | |
| Y-clamp | 3 Pi in a mer- geometry where the third Pi is distant to a pair of Pi representing the 10-membered-ring coordination |
| pdb id | IS Pi | IS Ribose | IS Other | OS Pi | OS Ribose | OS Base | Binding Free Energy (IS) kcal/mole | Binding Free Energy (OS) kcal/mole | Binding Free Energy (Total) kcal/mole |
|---|---|---|---|---|---|---|---|---|---|
| 4enc | A6, U7, U41, G42 | F− | −16.5 | 0 | −16.5 | ||||
| 3oxm | A33, C66 | G32 | U67, C68, U69 | −9.9 | −3.6 | −13.5 | |||
| 1hr2 | A171 | C165, U168, C170 | U167 | U167, A171 | −3.3 | −7.2 | −10.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cowan, J.A. Understanding the Thermodynamics of Magnesium Binding to RNA Structural Motifs. Life 2024, 14, 765. https://doi.org/10.3390/life14060765
Cowan JA. Understanding the Thermodynamics of Magnesium Binding to RNA Structural Motifs. Life. 2024; 14(6):765. https://doi.org/10.3390/life14060765
Chicago/Turabian StyleCowan, J. A. 2024. "Understanding the Thermodynamics of Magnesium Binding to RNA Structural Motifs" Life 14, no. 6: 765. https://doi.org/10.3390/life14060765
APA StyleCowan, J. A. (2024). Understanding the Thermodynamics of Magnesium Binding to RNA Structural Motifs. Life, 14(6), 765. https://doi.org/10.3390/life14060765






