Genetic Diversity of Trypanosoma cruzi in the United States of America: The Least Endemic Country for Chagas Disease
Abstract
1. Introduction
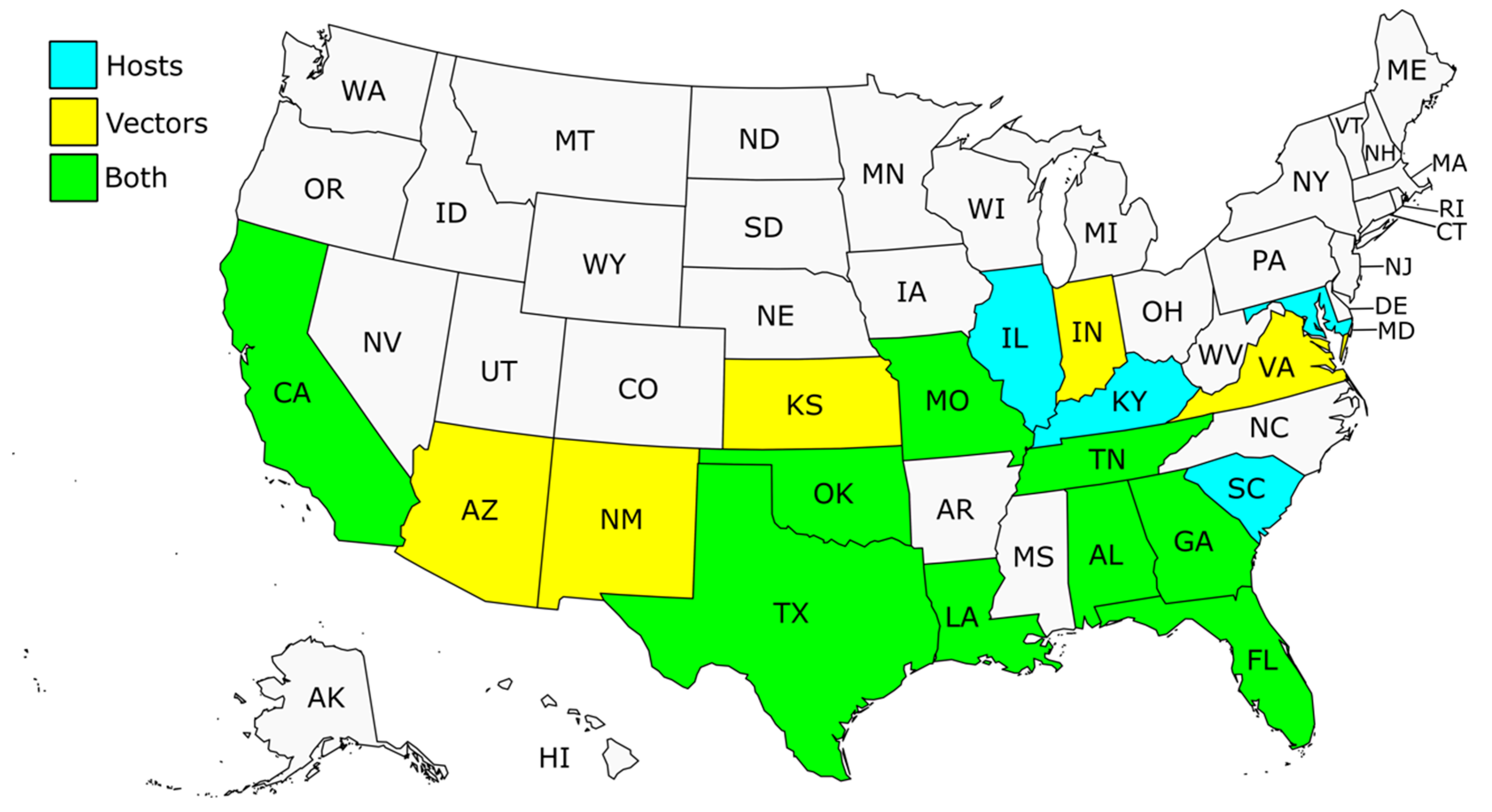
2. Chagas Disease Epidemiology in the US
Autochthonous Human Chagas Disease in the US
3. Trypanosoma cruzi Genetic Diversity
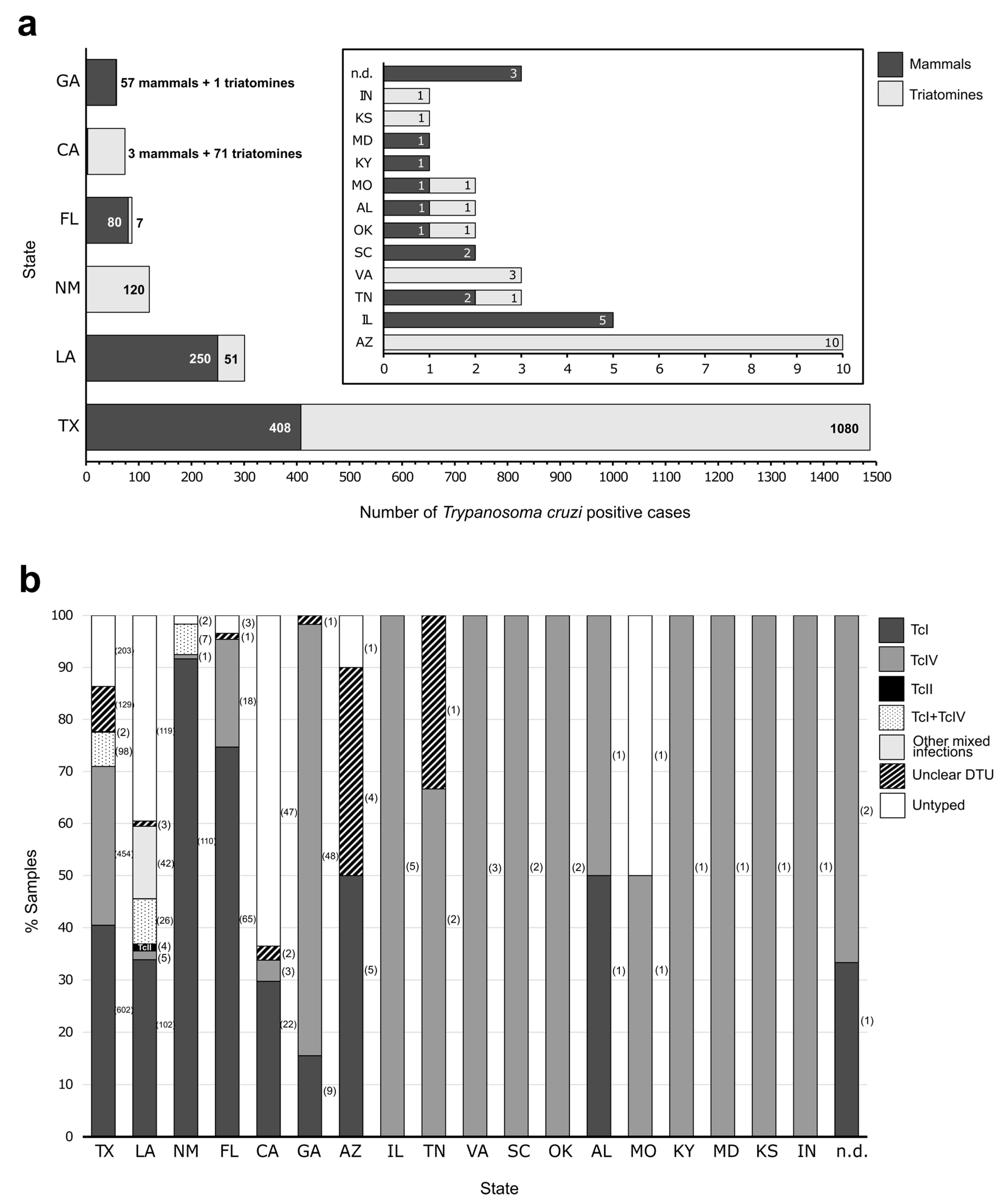
4. Trypanosoma cruzi DTUs in the US
4.1. Trypanosoma cruzi DTUs Identified in Hosts
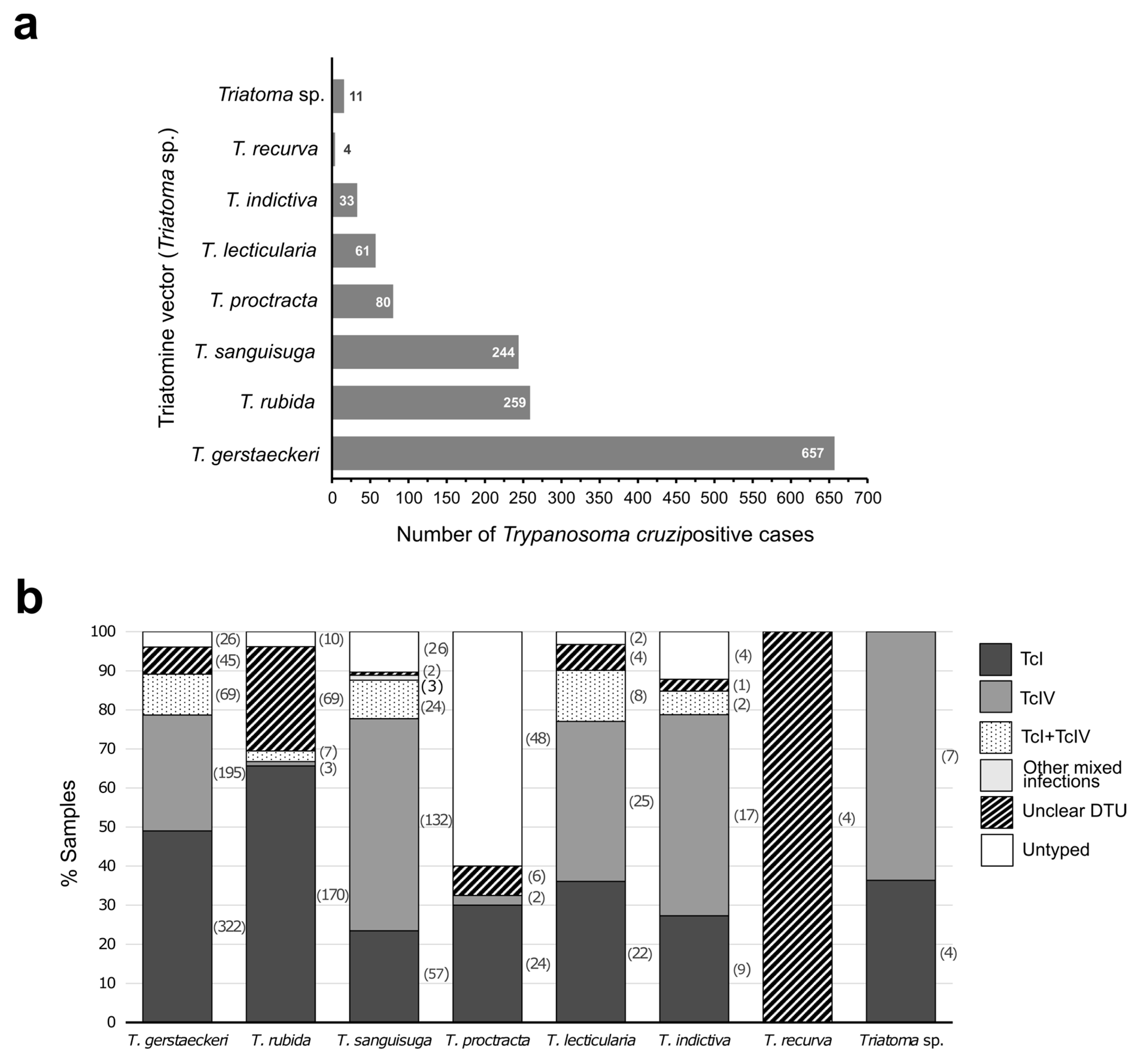
4.2. Trypanosoma cruzi DTUs Identified in Vectors
5. The Importance of Identifying DTUs in the US
6. The Problem of Identifying DTUs
7. Final Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Reference | Host | State | Target | Method |
Molecular Prevalence a P/Tested (%) |
Typed typ./P (%) | DTU (Number) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| TcI | TcIV | Other | Mixed Infections | |||||||
| Roellig et al. [32] | Human (Homo sapiens sapiens) | CA | SL-IR; 24Sα; 18S | PCR; MLEE; RAPD; STR | 2/2 (100) | 2/2 (100) | 2 | |||
| TX | 2/2 (100) | 2/2 (100) | 2 | |||||||
| LA | 1/1 (100) | 1/1 (100) | 1 | |||||||
| Domestic dog (Canis lupus familiaris) | n.d. | 2/2 (100) | 2/2 (100) | 2 * | ||||||
| TN | 1/1 (100) | 1/1 (100) | 1 TcI/TcIV * | |||||||
| OK | 1/1 (100) | 1/1 (100) | 1 * | |||||||
| SC | 1/1 (100) | 1/1 (100) | 1 * | |||||||
| CA | 1/1 (100) | 1/1 (100) | 1 * | |||||||
| Virginia opossum (Didelphis virginiana) | GA | 6/6 (100) | 6/6 (100) | 6 | ||||||
| FL | 6/6 (100) | 6/6 (100) | 6 | |||||||
| LA | 2/2 (100) | 2/2 (100) | 2 | |||||||
| AL | 1/1 (100) | 1/1 (100) | 1 | |||||||
| Raccoon (Procyon lotor) | GA | 45/45 (100) | 45/45 (100) | 2 | 43 * | |||||
| FL | 16/16 (100) | 16/16 (100) | 15 * | 1 TcI/TcIV * | ||||||
| SC | 1/1 (100) | 1/1 (100) | 1 * | |||||||
| MD | 1/1 (100) | 1/1 (100) | 1 * | |||||||
| TN | 1/1 (100) | 1/1 (100) | 1 * | |||||||
| Ring-tailed lemur (Lemur catta) | GA | 3/3 (100) | 3/3 (100) | 3 * | ||||||
| Rhesus macaque (Macaca mulatta) | n.d. | 1/1 (100) | 1/1 (100) | 1 | ||||||
| GA | 1/1 (100) | 1/1 (100) | 1 TcI/TcIV * | |||||||
| Nine-band armadillo (Dasypus novemcinctus) | GA | 1/1 (100) | 1/1 (100) | 1 * | ||||||
| LA | 2/2 (100) | 2/2 (100) | 2 | |||||||
| Striped skunk (Mephitis mephitis) | GA | 1/1 (100) | 1/1 (100) | 1 * | ||||||
| Charles et al. [48] | Southern plains woodrat (Neotoma micropus) | TX | 24Sα | PCR; sequencing | 35/104 (34) | 23/35 (66) | 10 | 13 | ||
| Striped skunks (M. mephitis) | TX | 4/4 (100) | 4/4 (100) | 1 | 3 | |||||
| Racoon (P. lotor) | TX | 12/20 (60) | 5/12 (42) | 5 | ||||||
| Hispid cotton rat (Sigmodon hispidus) | TX | 2/2 (100) | 2/2 (100) | 2 | ||||||
| Rock squirrel (Otospermophilus variegatus) | TX | 1/1 (100) | 1/1 (100) | 1 | ||||||
| Herrera et al. [49] | House mouse (Mus musculus); Cotton mouse (Peromyscus gossypinus) | LA | SL-IR; 24Sα; 18S | PCR; sequencing | 34/44 (77) | 20/34 (59) | 16 | 2 TcII | 1 TcI + TcII 1TcII + TcIV | |
| Eastern woodrat (Neotoma floridana) | LA | 11/15 (73) | 3/11 (27) | 2 | 1 TcII + TcIV | |||||
| Hodo et al. [50] | Evening bat (Nycticeius humeralis) | TX | SL-IR; 24Sα; 18S; COII | MTq-PCR | 1/593 (0.2) | 1/1 (100) | 1 | |||
| Curtis-Robles et al. [51] | Racoon (P. lotor) | TX | TcSC5D | Sequencing | 49/70 (70) | 11/49 (22) | 10 | 1 TcI + TcIV | ||
| Aleman et al. [52] | Northern pygmy mouse (Bayomis taylori) | TX | 18S | Sequencing | 1/4 (25) | 1/1 (100) | 1 | |||
| Southern plains woodrat (N. micropus) | TX | 1/5 (20) | 1/1 (100) | 1 | ||||||
| White-footed mouse (Peromyscus leucopus) | TX | 3/87 (3) | 3/3 (100) | 3 | ||||||
| Hispid cotton rat (Sigmodon hispidus) | TX | 1/13 (8) | 1/1 (100) | 1 | ||||||
| Hispid pocket mouse (Chaetodipus hispidus) | TX | 1/2 (50) | 1/1 (100) | 1 | ||||||
| Mexican spiny pocket mouse (Liomys irroratus) | TX | 1/44 (2) | 1/1 (100) | 1 | ||||||
| Curtis-Robles et al. [53] | Domestic dog (C. lupus familiaris) | TX | SL-IR; TcSC5D | MTq-PCR; sequencing | 15/86 (17) | 15/15 (100) | 9 | 5 | 1 TcI + TcIV | |
| Curtis-Robles et al. [54] | Domestic dog (C. lupus familiaris) | TX | SL-IR | MTq-PCR | 5/184 (3) | 4/5 (80) | 4 | |||
| Garcia et al. [31] | Human (H. sapiens sapiens) | TX | SL-IR; 24Sα; 18S | PCR; sequencing | 12/15 (80) | 6/12 (50) | 4 TcII/V/VI | 2 TcI + TcII/V/VI | ||
| Meyers et al. [55] | Domestic dog (C. lupus familiaris) | TX | SL-IR | MTq-PCR | 3/528 (0.6) | 2/3 (67) | 1 | 1 TcI + TcIV | ||
| Hodo et al. [56] | Rhesus macaque (M. mulatta) | TX | SL-IR; 24Sα; 18S; COII | MTq-PCR | 33/41 (80) | 33/33 (100) | 18 | 13 | 2 TcI + TcIV | |
| Virginia opossum (D. virginiana) | TX | 4/5 (80) | 4/4 (100) | 4 | ||||||
| Racoon (P. lotor) | TX | 2/5 (40) | 2/2 (100) | 2 | ||||||
| Striped skunk (M. mephitis) | TX | 2/3 (67) | 2/2 (100) | 1 | 1 | |||||
| Vandermark et al. [57] | Racoon (P. lotor) | IL | SL-IR; 24Sα | PCR; sequencing | 5 (37 global) | 5/5 (100) | 5 | |||
| Racoon (P. lotor) | KY | 1 (37 global) | 1/1 (100) | 1 | ||||||
| Racoon (P. lotor) | MO | 1 (37 global) | 1/1 (100) | 1 | ||||||
| Hodo et al. [58] | Domestic dog (C. lupus familiaris) | TX | SL-IR | MTq-PCR | 53/559 (9) | 6/53 (11) | 5 | 1 | ||
| Herrera et al. [59] | Rhesus macaque (M. mulatta) | LA | SL-IR | NGS; MB | 7/8 (88) | 7/7 (100) | 5 | 1 TcI + TcIV 1 TcI + TcVI | ||
| Pig-tailed macaque (Macaca nemestrina) | LA | 2/2 (100) | 2/2 (100) | 2 | ||||||
| Baboon (Papio spp.) | LA | 2/2 (100) | 2/2 (100) | 2 | ||||||
| Dumonteil et al. [60] | Domestic dog (C. lupus familiaris) | LA | SL-IR | PCR; NGS; MB | 73/540 (14) | 40/73 (55) | 25 | 10 TcI + TcIV 2 TcI + II + V + VI 2 TcI + II + IV + V + VI 1 TcI + TcII | ||
| Hodo et al. [61] | Coyote (Canis latrans) | TX | SL-IR; 24Sα; 18S; COII | MTq-PCR | 10/120 (8) | 10/10 (100) | 10 | |||
| Racoon (P. lotor) | TX | 15/24 (62) | 15/15 (100) | 15 | ||||||
| Meyers et al. [62] | Domestic dog (C. lupus familiaris) | TX | SL-IR | MTq-PCR | 4/1610 (0.2) | 3/4 (75) | 2 | 1 TcI + TcIV | ||
| Pronovost et al. [63] | House mouse (M. musculus) | LA | SL-IR | NGS; MB | 2/2 (100) | 2/2 (100) | 1 TcI + II + VI 1 TcII + VI | |||
| Cotton mouse (P. gossypinus) | LA | 3/3 (100) | 3/3 (100) | 1 TcII + TcVI 1 TcI + II + VI 1 TcI + II + V + VI | ||||||
| Eastern woodrat (N. floridana) | LA | 1/1 (100) | 1/1 (100) | 1 TcI + II + IV + VI | ||||||
| Zecca et al. [64] | Domestic cat (Felis catus) | TX | SL-IR; 24Sα; 18S; COII | MTq-PCR | 3/167 (2) | 3/3 (100) | 3 | |||
| Zecca et al. [65] | Virginia opossum (D. virginiana) | TX | SL-IR | MTq-PCR | 15/100 (15) | 15/15 (100) | 15 | |||
| Dumonteil et al. [66] | Domestic cat (Felis catus) | LA | SL-IR | PCR; sequencing | 70/284 (25) | 19/70 (27) | 3 ** | 16 ** | ||
| Rodríguez et al. [67] | Domestic dog (C. lupus familiaris) | TX | SL-IR; 24Sα; | PCR | 43/95 (45) | 40/43 (93) | 30 | 9 | 1 TcI + TcIV | |
| Domestic cat (Felis catus) | TX | 7/24 (42) | 3/7 (43) | 2 | 1 | |||||
| Cactus mouse (Peromyscus eremicus) | TX | 1/1 (100) | 1/1 (100) | 1 TcI + TcIV | ||||||
| Spotted ground squirrel (Xerospermophilus spilosoma) | TX | 1/1 (100) | 1/1 (100) | 1 TcI + TcIV | ||||||
| Western harvest mouse (Reithrodontomys megalotis) | TX | 1/1 (100) | 1/1 (100) | 1 | ||||||
| Gray fox (Urocyon cinereoargenteus) | TX | 1/1 (100) | 1/1 (100) | 1 | ||||||
| Coyote (C. latrans) | TX | 1/1 (100) | 1/1 (100) | 1 | ||||||
| Padilla et al. [68] | Cynomolgus macaque (Macaca fascicularis) | TX | LSU; SL-IR | PCR | 59/64 (92) | 40/59 (68) | 33 | 6 | 1 TcI + TcIV | |
| Torhorst et al. [69] | Virginia opossum (D. virginiana) | FL | SL-IR; 24Sα; 18S; COII | MTq-PCR | 58/112 (52) | 55/58 (95) | 55 | |||
| Majeau et al. [70] | Racoon (P. lotor) | LA | SL-IR | NGS | 40/119 (34) | 29/40 (73) | 19 | 2 TcII | 1 TcI + TcIV 7 *** | |
| Landsgaard et al. [71] | Domestic dog (C. lupus familiaris) | TX | SL-IR | PCR | 4/4 (100) | 3/4 (75) | 1 | 2 | ||
| Reference | Vector | State | Target | Method |
Molecular Prevalence a P/Tested (%) |
Typed typ./P (%) | DTU (Number) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| TcI | TcIV | Other | Mixed Infections | |||||||
| Roellig et al. [32] | Triatoma sanguisuga | FL | SL-IR; 24Sα; 18S | PCR; MLEE; RAPD; STR | 3/3 (100) | 3/3 (100) | 3 | |||
| T. sanguisuga | GA | 1/1 (100) | 1/1 (100) | 1 | ||||||
| Triatoma gerstaeckeri | TX | 3/3 (100) | 3/3 (100) | 2 | 1 * | |||||
| Cura et al. [45] | T. gerstaeckeri | TX | SL-IR | Sequencing | 7/7 (100) | 7/7 (100) | 7 | |||
| Hwang et al. [72] | Triatoma protracta | CA | 24Sα; 18S | Sequencing | 34/161 (21) | 2/34 (6) | 2 TcII/VI | |||
| Herrera et al. [49] | T. sanguisuga | LA | SL-IR; 24Sα; 18S | PCR; sequencing | 8/12 (67) | 6/8 (75) | 6 | |||
| Buhaya et al. [73] | Triatoma rubida | TX | TcSC5D | Sequencing | 25/39 (64) | 24/25 (96) | 24 | |||
| Shender et al. [74] | T. protracta | CA | SL-IR; 24Sα; HSP60; GPI | PCR; PCR-RFLP | 37/97 (38) | 22/37 (59) | 20 | 2 | ||
| Curtis-Robles et al. [53] | T. gerstaeckeri | TX | SL-IR; TcSC5D | MTq-PCR; sequencing | 16/16 (100) | 16/16 (100) | 10 | 4 | 2 TcI + TcIV | |
| T. sanguisuga | TX | 13/20 (65) | 13/13 (100) | 2 | 9 | 2 TcI + TcIV | ||||
| Curtis-Robles et al. [54] | T. gerstaeckeri | TX | SL-IR | MTq-PCR | 1/2 (50) | 1/1 (100) | 1 | |||
| Meyers et al. [55] | T. gerstaeckeri | TX | SL-IR | MTq-PCR | 9/18 (50) | 9/9 (100) | 6 | 1 | 2 TcI + TcIV | |
| Aleman et al. [52] | Triatoma lecticularia | TX | 18S | Sequencing | 13/19 (68) | 13/13 (100) | 13 | |||
| T. sanguisuga | TX | 2/2 (100) | 2/2 (100) | 2 | ||||||
| Curtis-Robles et al. [75] | T. gerstaeckeri | TX | SL-IR; TcSC5D | MTq-PCR; sequencing | 574/897 (64) | 548/574 (95) | 294 | 189 | 65 TcI + TcIV | |
| Triatoma indictiva | TX | 32/67 (48) | 28/32 (88) | 9 | 17 | 2 TcI + TcIV | ||||
| T. lecticularia | TX | 44/66 (67) | 42/44 (95) | 9 | 25 | 8 TcI + TcIV | ||||
| T. protracta | TX | 3/19 (16) | 2/3 (67) | 2 | ||||||
| T. rubida | TX | 11/64 (18) | 7/11 (64) | 6 | 1 | |||||
| T. sanguisuga | TX | 158/315 (50) | 135/158 (85) | 21 | 107 | 7 TcI + TcIV | ||||
| Triatoma sp. | TX | 11/29 (38) | 11/11 (100) | 4 | 7 | |||||
| T. sanguisuga | AL | 1/4 (25) | 1/1 (100) | 1 | ||||||
| T. protracta | AZ | 1/7 (14) | 1/1 (100) | 1 | ||||||
| T. rubida | AZ | 5/33 (15) | 4/5 (80) | 4 | ||||||
| T. sanguisuga | FL | 4/25 (16) | 4/4 (100) | 1 | 3 | |||||
| T. sanguisuga | IN | 1/2 (50) | 1/1 (100) | 1 | ||||||
| T. sanguisuga | KS | 1/1 (100) | 1/1 (100) | 1 | ||||||
| T. sanguisuga | LA | 3/3 (100) | 3/3 (100) | 1 | 2 | |||||
| T. sanguisuga | MO | 1/2 (50) | 0/1 (0) | |||||||
| T. rubida | NM | 2/7 (29) | 1/2 (100) | 1 | ||||||
| T. sanguisuga | OK | 1/2 (50) | 1/1 (100) | 1 | ||||||
| T. sanguisuga | TN | 1/2 (50) | 1/1 (100) | 1 | ||||||
| T. sanguisuga | VA | 3/7 (43) | 3/3 (100) | 3 | ||||||
| Curtis-Robles et al. [76] | T. gerstaeckeri | TX | SL-IR | MTq-PCR | 1/11 (9) | n.s. | n.s. ** | |||
| T. protracta | TX | 4/9 (44) | n.s. | n.s. ** | ||||||
| T. rubida | TX | 69/299 (23) | n.s. | n.s. ** | ||||||
| Hodo et al. [56] | T. sanguisuga | TX | SL-IR; 24Sα; 18S; COII | MTq-PCR | 1/4 (25) | 1/1 (100) | 1 | |||
| Dumonteil et al. [77] | T. sanguisuga | LA | SL-IR | MTq-PCR; sequencing | 40/45 (89) | 40/40 (100) | 19 | 3 | 15 TcI + TcIV 2 TcI + TcII/V 1 TcI + IV + II/V | |
| Rodríguez et al. [67] | T. rubida | TX | SL-IR; 24Sα; | PCR | 29/50 (58) | 27/29 (93) | 26 | 1 | ||
| T. protracta | TX | 1/2 (50) | 1/1 (100) | 1 | ||||||
| T. gerstaeckeri | TX | 2/2 (100) | 2/2 (100) | 2 | ||||||
| T. rubida | NM | 118/171 (69) | 117/118 (99) | 109 | 1 | 7 TcI + TcIV | ||||
| Flores-López et al. [78] | T. gerstaeckeri | TX | COII-ND1; MSH2; DHFR-TS; TcCLB | MLST | 44/n.s. | n.s. *** | 75% *** | 25% *** | ||
| T. lecticularia | TX | 4/n.s. | ||||||||
| T. indictiva | TX | 1/n.s. | ||||||||
| T. sanguisuga | TX | 2/n.s. | ||||||||
| Triatoma recurva | AZ | 4/n.s. | ||||||||
References
- Rassi, A., Jr.; Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Noireau, F.; Diosque, P.; Jansen, A.M. Trypanosoma cruzi: Adaptation to its vectors and its hosts. Vet. Res. 2009, 40, 26. [Google Scholar] [CrossRef]
- Bern, C.; Kjos, S.; Yabsley, M.J.; Montgomery, S.P. Trypanosoma cruzi and Chagas’ Disease in the United States. Clin. Microbiol. Rev. 2011, 24, 655–681. [Google Scholar] [CrossRef]
- Coura, J.R.; Viñas, P.A.; Junqueira, A.C. Ecoepidemiology, short history and control of Chagas disease in the endemic countries and the new challenge for non-endemic countries. Mem. Inst. Oswaldo Cruz 2014, 109, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Justi, S.A.; Galvão, C. The Evolutionary Origin of Diversity in Chagas Disease Vectors. Trends Parasitol. 2017, 33, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Coura, J.R.; Dias, J.C. Epidemiology, control and surveillance of Chagas disease: 100 years after its discovery. Mem. Inst. Oswaldo Cruz 2009, 104 (Suppl. S1), 31–40. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, C.M.; Cavalcanti, L.P.; Souza, R.C.; Barbosa, S.E.; Xavier, S.C.; Jansen, A.M.; Ramalho, R.D.; Diotaiut, L. Domestic, peridomestic and wild hosts in the transmission of Trypanosoma cruzi in the Caatinga area colonised by Triatoma brasiliensis. Mem. Inst. Oswaldo Cruz 2014, 109, 887–898. [Google Scholar] [CrossRef]
- Gourbière, S.; Dorn, P.; Tripet, F.; Dumonteil, E. Genetics and evolution of triatomines: From phylogeny to vector control. Heredity 2012, 108, 190–202. [Google Scholar] [CrossRef]
- Velásquez-Ortiz, N.; Ramírez, J.D. Understanding the oral transmission of Trypanosoma cruzi as a veterinary and medical foodborne zoonosis. Res. Vet. Sci. 2020, 132, 448–461. [Google Scholar] [CrossRef]
- Pérez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- De Sousa, A.S.; Vermeij, D.; Ramos, A.N., Jr.; Luquetti, A.O. Chagas disease. Lancet 2023, 403, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Abras, A.; Ballart, C.; Fernández-Arévalo, A.; Pinazo, M.J.; Gascón, J.; Muñoz, C.; Gállego, M. Worldwide Control and Management of Chagas Disease in a New Era of Globalization: A Close Look at Congenital Trypanosoma cruzi Infection. Clin. Microbiol. Rev. 2022, 35, e0015221. [Google Scholar] [CrossRef]
- Pan American Health Organization (PAHO). Chagas Disease in the Americas: An Analysis of the Current Situation and Strategic Review of the Regional Agenda. Final Report, 14–16 March 2023, Medellín, Colombia; PAHO: Washington, DC, USA, 2023. [Google Scholar]
- Gascón, J.; Bern, C.; Pinazo, M.J. Chagas disease in Spain, the United States and other non-endemic countries. Acta Trop. 2010, 115, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Albajar-Vinas, P.; Jannin, J. The hidden Chagas disease burden in Europe. Euro Surveill. 2011, 16, 19975. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gascón, J.; Requena Méndez, A.; Pinazo Delgado, M.-J. Enfermedad de Chagas en países no endémicos. In Enfermedad de Chagas. Un Enfoque Práctico Basado en la Investigación Médica, 1st ed.; Viotti, R.J., Vigliano, C.A., Eds.; Médica Panamericana: Buenos Aires, Argentina, 2015; Volume 1, pp. 56–67. [Google Scholar]
- Forsyth, C.J.; Manne-Goehler, J.; Bern, C.; Whitman, J.; Hochberg, N.S.; Edwards, M.; Marcus, R.; Beatty, N.L.; Castro-Sesquen, Y.E.; Coyle, C.; et al. Recommendations for Screening and Diagnosis of Chagas Disease in the United States. J. Infect. Dis. 2022, 225, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Bern, C.; Messenger, L.A.; Whitman, J.D.; Maguire, J.H. Chagas Disease in the United States: A Public Health Approach. Clin. Microbiol. Rev. 2020, 33, e00023-19. [Google Scholar] [CrossRef]
- Abras, A.; Gállego, M.; Muñoz, C.; Juiz, N.A.; Ramírez, J.C.; Cura, C.I.; Tebar, S.; Fernández-Arévalo, A.; Pinazo, M.J.; de la Torre, L.; et al. Identification of Trypanosoma cruzi Discrete Typing Units (DTUs) in Latin-American migrants in Barcelona (Spain). Parasitol. Int. 2017, 66, 83–88. [Google Scholar] [CrossRef]
- Cura, C.I.; Schijman, A.G. Relación entre los genotipados de T. cruzi y la presentación clínica de la enfermedad de Chagas. Rev. Esp. Salud Pública 2013, 86, 9–16. [Google Scholar]
- Zingales, B.; Macedo, A.M. Fifteen Years after the Definition of Trypanosoma cruzi DTUs: What Have We Learned? Life 2023, 13, 2339. [Google Scholar] [CrossRef]
- Lynn, M.K.; Bossak, B.H.; Sandifer, P.A.; Watson, A.; Nolan, M.S. Contemporary autochthonous human Chagas disease in the USA. Acta Trop. 2020, 205, 105361. [Google Scholar] [CrossRef]
- Garza, M.; Feria Arroyo, T.P.; Casillas, E.A.; Sanchez-Cordero, V.; Rivaldi, C.L.; Sarkar, S. Projected future distributions of vectors of Trypanosoma cruzi in North America under climate change scenarios. PLoS Negl. Trop. Dis. 2014, 8, e2818. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Castro, O.; Moo-Llanes, D.A.; Ramsey, J.M. Impact of climate change on vector transmission of Trypanosoma cruzi (Chagas, 1909) in North America. Med. Vet. Entomol. 2018, 32, 84–101. [Google Scholar] [CrossRef]
- Manne-Goehler, J.; Umeh, C.A.; Montgomery, S.P.; Wirtz, V.J. Estimating the Burden of Chagas Disease in the United States. PLoS Negl. Trop. Dis. 2016, 10, e0005033. [Google Scholar] [CrossRef]
- Manne-Goehler, J.; Reich, M.R.; Wirtz, V.J. Access to care for Chagas disease in the United States: A health systems analysis. Am. J. Trop. Med. Hyg. 2015, 93, 108–113. [Google Scholar] [CrossRef]
- Edwards, M.S.; Stimpert, K.K.; Bialek, S.R.; Montgomery, S.P. Evaluation and Management of Congenital Chagas Disease in the United States. J. Pediatric. Infect. Dis. Soc. 2019, 8, 461–469. [Google Scholar] [CrossRef]
- Montgomery, S.P.; Parise, M.E.; Dotson, E.M.; Bialek, S.R. What Do We Know About Chagas Disease in the United States? Am. J. Trop. Med. Hyg. 2016, 95, 1225–1227. [Google Scholar] [CrossRef]
- Turabelidze, G.; Vasudevan, A.; Rojas-Moreno, C.; Montgomery, S.P.; Baker, M.; Pratt, D.; Enyeart, S. Autochthonous Chagas Disease–Missouri, 2018. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 193–195. [Google Scholar] [CrossRef]
- Lynn, M.K.; Dye-Braumuller, K.C.; Beatty, N.L.; Dorn, P.L.; Klotz, S.A.; Stramer, S.L.; Townsend, R.L.; Kamel, H.; Vannoy, J.M.; Sadler, P.; et al. Evidence of likely autochthonous Chagas disease in the southwestern United States: A case series of Trypanosoma cruzi seropositive blood donors. Transfusion 2022, 62, 1808–1817. [Google Scholar] [CrossRef]
- Garcia, M.N.; Burroughs, H.; Gorchakov, R.; Gunter, S.M.; Dumonteil, E.; Murray, K.O.; Herrera, C.P. Molecular identification and genotyping of Trypanosoma cruzi DNA in autochthonous Chagas disease patients from Texas, USA. Infect. Genet. Evol. 2017, 49, 151–156. [Google Scholar] [CrossRef]
- Roellig, D.M.; Brown, E.L.; Barnabé, C.; Tibayrenc, M.; Steurer, F.J.; Yabsley, M.J. Molecular typing of Trypanosoma cruzi isolates, United States. Emerg. Infect. Dis. 2008, 14, 1123–1125. [Google Scholar] [CrossRef]
- Beatty, N.L.; Klotz, S.A. Autochthonous Chagas Disease in the United States: How Are People Getting Infected? Am. J. Trop. Med. Hyg. 2020, 103, 967–969. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002, 8, 405–410. [Google Scholar] [CrossRef]
- Berry, A.S.F.; Salazar-Sánchez, R.; Castillo-Neyra, R.; Borrini-Mayorí, K.; Chipana-Ramos, C.; Vargas-Maquera, M.; Ancca-Juarez, J.; Náquira-Velarde, C.; Levy, M.Z.; Brisson, D.; et al. Sexual reproduction in a natural Trypanosoma cruzi population. PLoS Negl. Trop. Dis. 2019, 13, e0007392. [Google Scholar] [CrossRef]
- Schwabl, P.; Imamura, H.; Van den Broeck, F.; Costales, J.A.; Maiguashca-Sánchez, J.; Miles, M.A.; Andersson, B.; Grijalva, M.J.; Llewellyn, M.S. Meiotic sex in Chagas disease parasite Trypanosoma cruzi. Nat. Commun. 2019, 10, 3972. [Google Scholar] [CrossRef]
- Tibayrenc, M. Genetic epidemiology of parasitic protozoa and other infectious agents: The need for an integrated approach. Int. J. Parasitol. 1998, 8, 85–104. [Google Scholar] [CrossRef]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef]
- Anonymous. Recommendations from a satellite meeting. Mem. Inst. Oswaldo Cruz 1999, 1, 429–432. [Google Scholar]
- Brisse, S.; Dujardin, J.C.; Tibayrenc, M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol. Biochem. Parasitol. 2000, 111, 95–105. [Google Scholar] [CrossRef]
- Brisse, S.; Verhoef, J.; Tibayrenc, M. Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int. J. Parasitol. 2001, 31, 1218–1226. [Google Scholar] [CrossRef]
- Zingales, B.; Andrade, S.G.; Briones, M.R.; Campbell, D.A.; Chiari, E.; Fernandes, O.; Guhl, F.; Lages-Silva, E.; Macedo, A.M.; Machado, C.R.; et al. Second Satellite Meeting. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz 2009, 104, 1051–1054. [Google Scholar] [CrossRef]
- Zingales, B.; Bartholomeu, D.C. Trypanosoma cruzi genetic diversity: Impact on transmission cycles and Chagas disease. Mem. Inst. Oswaldo Cruz 2022, 117, e210193. [Google Scholar] [CrossRef]
- Guhl, F.; Ramírez, J.D. Trypanosoma cruzi I diversity: Towards the need of genetic subdivision? Acta Trop. 2011, 119, 1–4. [Google Scholar] [CrossRef]
- Cura, C.I.; Mejía-Jaramillo, A.M.; Duffy, T.; Burgos, J.M.; Rodriguero, M.; Cardinal, M.V.; Kjos, S.; Gurgel-Gonçalves, R.; Blanchet, D.; De Pablos, L.M.; et al. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int. J. Parasitol. 2010, 40, 1599–1607. [Google Scholar] [CrossRef]
- Marcili, A.; Lima, L.; Cavazzana, M.; Junqueira, A.C.; Veludo, H.H.; Maia Da Silva, F.; Campaner, M.; Paiva, F.; Nunes, V.L.; Teixeira, M.M. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology 2009, 136, 641–655. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Hernández, C.; Montilla, M.; Zambrano, P.; Flórez, A.C.; Parra, E.; Cucunubá, Z.M. First report of human Trypanosoma cruzi infection attributed to TcBat genotype. Zoonoses Public Health 2014, 61, 477–479. [Google Scholar] [CrossRef]
- Charles, R.A.; Kjos, S.; Ellis, A.E.; Barnes, J.C.; Yabsley, M.J. Southern plains woodrats (Neotoma micropus) from southern Texas are important reservoirs of two genotypes of Trypanosoma cruzi and host of a putative novel Trypanosoma species. Vector Borne Zoonotic Dis. 2013, 13, 22–30. [Google Scholar] [CrossRef]
- Herrera, C.P.; Licon, M.H.; Nation, C.S.; Jameson, S.B.; Wesson, D.M. Genotype diversity of Trypanosoma cruzi in small rodents and Triatoma sanguisuga from a rural area in New Orleans, Louisiana. Parasit. Vectors 2015, 8, 123. [Google Scholar] [CrossRef]
- Hodo, C.L.; Goodwin, C.C.; Mayes, B.C.; Mariscal, J.A.; Waldrup, K.A.; Hamer, S.A. Trypanosome species, including Trypanosoma cruzi, in sylvatic and peridomestic bats of Texas, USA. Acta Trop. 2016, 164, 259–266. [Google Scholar] [CrossRef]
- Curtis-Robles, R.; Lewis, B.C.; Hamer, S.A. High Trypanosoma cruzi infection prevalence associated with minimal cardiac pathology among wild carnivores in central Texas. Int. J. Parasitol. Parasites Wildl. 2016, 5, 117–123. [Google Scholar] [CrossRef]
- Aleman, A.; Guerra, T.; Maikis, T.J.; Milholland, M.T.; Castro-Arellano, I.; Forstner, M.R.J.; Hahn, D. The Prevalence of Trypanosoma cruzi, the Causal Agent of Chagas Disease, in Texas Rodent Populations. Ecohealth 2017, 14, 130–143. [Google Scholar] [CrossRef]
- Curtis-Robles, R.; Snowden, K.F.; Dominguez, B.; Dinges, L.; Rodgers, S.; Mays, G.; Hamer, S.A. Epidemiology and Molecular Typing of Trypanosoma cruzi in Naturally-Infected Hound Dogs and Associated Triatomine Vectors in Texas, USA. PLoS Negl. Trop. Dis. 2017, 11, e0005298. [Google Scholar] [CrossRef]
- Curtis-Robles, R.; Zecca, I.B.; Roman-Cruz, V.; Carbajal, E.S.; Auckland, L.D.; Flores, I.; Millard, A.V.; Hamer, S.A. Trypanosoma cruzi (Agent of Chagas Disease) in Sympatric Human and Dog Populations in “Colonias” of the Lower Rio Grande Valley of Texas. Am. J. Trop. Med. Hyg. 2017, 96, 805–814. [Google Scholar] [CrossRef]
- Meyers, A.C.; Meinders, M.; Hamer, S.A. Widespread Trypanosoma cruzi infection in government working dogs along the Texas-Mexico border: Discordant serology, parasite genotyping and associated vectors. PLoS Negl. Trop. Dis. 2017, 11, e0005819. [Google Scholar] [CrossRef]
- Hodo, C.L.; Wilkerson, G.K.; Birkner, E.C.; Gray, S.B.; Hamer, S.A. Trypanosoma cruzi Transmission Among Captive Nonhuman Primates, Wildlife, and Vectors. EcoHealth 2018, 15, 426–436. [Google Scholar] [CrossRef]
- Vandermark, C.; Zieman, E.; Boyles, E.; Nielsen, C.K.; Davis, C.; Jiménez, F.A. Trypanosoma cruzi strain TcIV infects raccoons from Illinois. Mem. Inst. Oswaldo Cruz 2018, 113, 30–37. [Google Scholar] [CrossRef]
- Hodo, C.L.; Rodriguez, J.Y.; Curtis-Robles, R.; Zecca, I.B.; Snowden, K.F.; Cummings, K.J.; Hamer, S.A. Repeated cross-sectional study of Trypanosoma cruzi in shelter dogs in Texas, in the context of Dirofilaria immitis and tick-borne pathogen prevalence. J. Vet. Intern. Med. 2019, 33, 158–166. [Google Scholar] [CrossRef]
- Herrera, C.; Majeau, A.; Didier, P.; Falkenstein, K.P.; Dumonteil, E. Trypanosoma cruzi diversity in naturally infected nonhuman primates in Louisiana assessed by deep sequencing of the mini-exon gene. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 281–286. [Google Scholar] [CrossRef]
- Dumonteil, E.; Elmayan, A.; Majeau, A.; Tu, W.; Duhon, B.; Marx, P.; Wolfson, W.; Balsamo, G.; Herrera, C. Genetic diversity of Trypanosoma cruzi parasites infecting dogs in southern Louisiana sheds light on parasite transmission cycles and serological diagnostic performance. PLoS Negl. Trop. Dis. 2020, 14, e0008932. [Google Scholar] [CrossRef]
- Hodo, C.L.; Bañuelos, R.M.; Edwards, E.E.; Wozniak, E.J.; Hamer, S.A. Pathology and discrete typing unit associations of Trypanosoma cruzi infection in coyotes (Canis latrans) and raccoons (Procyon lotor) of Texas, USA. J. Wildl. Dis. 2020, 56, 134–144. [Google Scholar] [CrossRef]
- Meyers, A.C.; Purnell, J.C.; Ellis, M.M.; Auckland, L.D.; Meinders, M.; Hamer, S.A. Nationwide Exposure of U.S. Working Dogs to the Chagas Disease Parasite, Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 2020, 102, 1078–1085. [Google Scholar] [CrossRef]
- Pronovost, H.; Peterson, A.C.; Chavez, B.G.; Blum, M.J.; Dumonteil, E.; Herrera, C.P. Deep sequencing reveals multiclonality and new discrete typing units of Trypanosoma cruzi in rodents from the southern United States. J. Microbiol. Immunol. Infect. 2020, 53, 622–633. [Google Scholar] [CrossRef]
- Zecca, I.B.; Hodo, C.L.; Slack, S.; Auckland, L.; Rodgers, S.; Killets, K.C.; Saunders, A.B.; Hamer, S.A. Prevalence of Trypanosoma cruzi infection and associated histologic findings in domestic cats (Felis catus). Vet. Parasitol. 2020, 278, 109014. [Google Scholar] [CrossRef]
- Zecca, I.B.; Hodo, C.L.; Slack, S.; Auckland, L.; Hamer, S.A. Trypanosoma cruzi infections and associated pathology in urban-dwelling Virginia opossums (Didelphis virginiana). Int. J. Parasitol. Parasites Wildl. 2020, 11, 287–293. [Google Scholar] [CrossRef]
- Dumonteil, E.; Desale, H.; Tu, W.; Duhon, B.; Wolfson, W.; Balsamo, G.; Herrera, C. Shelter cats host infections with multiple Trypanosoma cruzi discrete typing units in southern Louisiana. Vet. Res. 2021, 52, 53. [Google Scholar] [CrossRef]
- Rodriguez, F.; Luna, B.S.; Calderon, O.; Manriquez-Roman, C.; Amezcua-Winter, K.; Cedillo, J.; Garcia-Vazquez, R.; Tejeda, I.A.; Romero, A.; Waldrup, K.; et al. Surveillance of Trypanosoma cruzi infection in Triatomine vectors, feral dogs and cats, and wild animals in and around El Paso county, Texas, and New Mexico. PLoS Negl. Trop. Dis. 2021, 15, e0009147. [Google Scholar] [CrossRef]
- Padilla, A.M.; Yao, P.Y.; Landry, T.J.; Cooley, G.M.; Mahaney, S.M.; Ribeiro, I.; VandeBerg, J.L.; Tarleton, R.L. High variation in immune responses and parasite phenotypes in naturally acquired Trypanosoma cruzi infection in a captive non-human primate breeding colony in Texas, USA. PLoS Negl. Trop. Dis. 2021, 15, e0009141. [Google Scholar] [CrossRef]
- Torhorst, C.W.; White, Z.S.; Bhosale, C.R.; Beatty, N.L.; Wisely, S.M. Identification of the parasite, Trypanosoma cruzi, in multiple tissues of epidemiological significance in the Virginia opossum (Didelphis virginiana): Implications for environmental and vertical transmission routes. PLoS Negl. Trop. Dis. 2022, 16, e0010974. [Google Scholar] [CrossRef]
- Majeau, A.; Cloherty, E.; Anderson, A.N.; Straif-Bourgeois, S.C.; Dumonteil, E.; Herrera, C. Genetic diversity of Trypanosoma cruzi infecting raccoons (Procyon lotor) in 2 metropolitan areas of southern Louisiana: Implications for parasite transmission networks. Parasitology 2023, 150, 374–381. [Google Scholar] [CrossRef]
- Landsgaard, K.A.; Milliron, S.M.; Faccin, M.; Broughton, C.A.; Auckland, L.D.; Edwards, J.F.; Hamer, S.A.; Hensel, M.E. Protozoal meningoencephalitis and myelitis in 4 dogs associated with Trypanosoma cruzi infection. Vet. Pathol. 2023, 60, 199–202. [Google Scholar] [CrossRef]
- Hwang, W.S.; Zhang, G.; Maslov, D.; Weirauch, C. Infection rates of Triatoma protracta (Uhler) with Trypanosoma cruzi in Southern California and molecular identification of trypanosomes. Am. J. Trop. Med. Hyg. 2010, 83, 1020–1022. [Google Scholar] [CrossRef]
- Buhaya, M.H.; Galvan, S.; Maldonado, R.A. Incidence of Trypanosoma cruzi infection in triatomines collected at Indio Mountains Research Station. Acta Trop. 2015, 150, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Shender, L.A.; Lewis, M.D.; Rejmanek, D.; Mazet, J.A. Molecular Diversity of Trypanosoma cruzi Detected in the Vector Triatoma protracta from California, USA. PLoS Negl. Trop. Dis. 2016, 10, e0004291. [Google Scholar] [CrossRef] [PubMed]
- Curtis-Robles, R.; Auckland, L.D.; Snowden, K.F.; Hamer, G.L.; Hamer, S.A. Analysis of over 1500 triatomine vectors from across the US, predominantly Texas, for Trypanosoma cruzi infection and discrete typing units. Infect. Genet. Evol. 2018, 58, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Curtis-Robles, R.; Meyers, A.C.; Auckland, L.D.; Zecca, I.B.; Skiles, R.; Hamer, S.A. Parasitic interactions among Trypanosoma cruzi, triatomine vectors, domestic animals, and wildlife in Big Bend National Park along the Texas-Mexico border. Acta Trop. 2018, 188, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Dumonteil, E.; Pronovost, H.; Bierman, E.F.; Sanford, A.; Majeau, A.; Moore, R.; Herrera, C. Interactions among Triatoma sanguisuga blood feeding sources, gut microbiota and Trypanosoma cruzi diversity in southern Louisiana. Mol Ecol. 2020, 29, 3747–3761. [Google Scholar] [CrossRef] [PubMed]
- Flores-López, C.A.; Mitchell, E.A.; Reisenman, C.E.; Sarkar, S.; Williamson, P.C.; Machado, C.A. Phylogenetic diversity of two common Trypanosoma cruzi lineages in the Southwestern United States. Infect. Genet. Evol. 2022, 99, 105251. [Google Scholar] [CrossRef] [PubMed]
- Barnabé, C.; De Meeûs, T.; Noireau, F.; Bosseno, M.F.; Monje, E.M.; Renaud, F.; Brenière, S.F. Trypanosoma cruzi discrete typing units (DTUs): Microsatellite loci and population genetics of DTUs TcV and TcI in Bolivia and Peru. Infect. Genet. Evol 2011, 11, 1752–1760. [Google Scholar] [CrossRef]
- Majeau, A.; Pronovost, H.; Sanford, A.; Cloherty, E.; Anderson, A.N.; Balsamo, G.; Gee, L.; Straif-Bourgeois, S.C.; Herrera, C. Raccoons As an Important Reservoir for Trypanosoma cruzi: A Prevalence Study from Two Metropolitan Areas in Louisiana. Vector Borne Zoonotic Dis. 2020, 20, 535–540. [Google Scholar] [CrossRef]
- Klotz, S.A.; Dorn, P.L.; Mosbacher, M.; Schmidt, J.O. Kissing bugs in the United States: Risk for vector-borne disease in humans. Environ. Health Insights 2014, 8 (Suppl. S2), 49–59. [Google Scholar] [CrossRef]
- Monje-Rumi, M.M.; Floridia-Yapur, N.; Zago, M.P.; Ragone, P.G.; Pérez Brandán, C.M.; Nuñez, S.; Barrientos, N.; Tomasini, N.; Diosque, P. Potential association of Trypanosoma cruzi DTUs TcV and TcVI with the digestive form of Chagas disease. Infect. Genet. Evol. 2020, 84, 104329. [Google Scholar] [CrossRef]
- Majeau, A.; Murphy, L.; Herrera, C.; Dumonteil, E. Assessing Trypanosoma cruzi Parasite Diversity through Comparative Genomics: Implications for Disease Epidemiology and Diagnostics. Pathogens 2021, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Prates Nielebock, M.A.; Moreira, O.C.; das Chagas Xavier, S.C.; Miranda, L.F.C.; Bastos de Lima, A.C.; Oliveira de Jesus Sales Pereira, T.; Hasslocher-Moreno, A.M.; Britto, C.; Conde Sangenis, L.H.; Magalhães Saraiva, R. Association between Trypanosoma cruzi DTU TcII and chronic Chagas disease clinical presentation and outcome in an urban cohort in Brazil. PLoS ONE 2020, 15, e0243008. [Google Scholar]
- Magalhães, L.M.D.; Gollob, K.J.; Zingales, B.; Dutra, W.O. Pathogen diversity, immunity, and the fate of infections: Lessons learned from Trypanosoma cruzi human-host interactions. Lancet Microbe 2022, 3, e711–e722. [Google Scholar] [CrossRef] [PubMed]
- Carlier, Y.; Sosa-Estani, S.; Luquetti, A.O.; Buekens, P. Congenital Chagas disease: An update. Mem. Inst. Oswaldo Cruz 2015, 110, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Ayres, J.; Marcus, R.; Standley, C.J. The Importance of Screening for Chagas Disease against the Backdrop of Changing Epidemiology in the USA. Curr. Trop. Med. 2022, 9, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J. The rise of neglected tropical diseases in the “new Texas”. PLoS Negl. Trop. Dis. 2018, 12, e0005581. [Google Scholar] [CrossRef] [PubMed]
- Silvestrini, M.M.A.; Alessio, G.D.; Frias, B.E.D.; Sales Júnior, P.A.; Araújo, M.S.S.; Silvestrini, C.M.A.; Brito Alvim de Melo, G.E.; Martins-Filho, O.A.; Teixeira-Carvalho, A.; Martins, H.R. New insights into Trypanosoma cruzi genetic diversity, and its influence on parasite biology and clinical outcomes. Front. Immunol. 2024, 15, 1342431. [Google Scholar] [CrossRef] [PubMed]
- Meymandi, S.; Hernandez, S.; Park, S.; Sanchez, D.R.; Forsyth, C. Treatment of Chagas Disease in the United States. Curr. Treat. Options Infect. Dis. 2018, 10, 373–388. [Google Scholar] [CrossRef]
- Jiménez, P.; Jaimes, J.; Poveda, C.; Ramírez, J.D. A systematic review of the Trypanosoma cruzi genetic heterogeneity, host immune response and genetic factors as plausible drivers of chronic chagasic cardiomyopathy. Parasitology 2019, 146, 269–283. [Google Scholar] [CrossRef]
- Vela, A.; Coral-Almeida, M.; Sereno, D.; Costales, J.A.; Barnabé, C.; Brenière, S.F. In vitro susceptibility of Trypanosoma cruzi discrete typing units (DTUs) to benznidazole: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2021, 15, e0009269. [Google Scholar] [CrossRef]
- Revollo, S.; Oury, B.; Vela, A.; Tibayrenc, M.; Sereno, D. In Vitro Benznidazole and Nifurtimox Susceptibility Profile of Trypanosoma cruzi Strains Belonging to Discrete Typing Units TcI, TcII, and TcV. Pathogens 2019, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Agudelo Higuita, N.I.; Beatty, N.L.; Forsyth, C.; Henao-Martínez, A.F.; Manne-Goehler, J.; US Chagas Research Consortium. Chagas disease in the United States: A call for increased investment and collaborative research. Lancet Reg. Health Am. 2024, 34, 100768. [Google Scholar] [CrossRef] [PubMed]
- Burgos, J.M.; Diez, M.; Vigliano, C.; Bisio, M.; Risso, M.; Duffy, T.; Cura, C.; Brusses, B.; Favaloro, L.; Leguizamon, M.S.; et al. Molecular identification of Trypanosoma cruzi discrete typing units in end-stage chronic Chagas heart disease and reactivation after heart transplantation. Clin. Infect. Dis. 2010, 51, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Cura, C.I.; Duffy, T.; Lucero, R.H.; Bisio, M.; Péneau, J.; Jimenez-Coello, M.; Calabuig, E.; Gimenez, M.J.; Valencia Ayala, E.; Kjos, S.A.; et al. Multiplex Real-Time PCR Assay Using TaqMan Probes for the Identification of Trypanosoma cruzi DTUs in Biological and Clinical Samples. PLoS Negl. Trop. Dis. 2015, 9, e0003765. [Google Scholar] [CrossRef] [PubMed]
- Velásquez-Ortiz, N.; Herrera, G.; Hernández, C.; Muñoz, M.; Ramírez, J.D. Discrete typing units of Trypanosoma cruzi: Geographical and biological distribution in the Americas. Sci. Data 2022, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.C.; Torres, C.; Curto, M.L.A.; Schijman, A.G. New insights into Trypanosoma cruzi evolution, genotyping and molecular diagnostics from satellite DNA sequence analysis. PLoS Negl. Trop. Dis. 2017, 11, e0006139. [Google Scholar] [CrossRef] [PubMed]
- Rusman, F.; Tomasini, N.; Yapur, N.F.; Puebla, A.F.; Ragone, P.G.; Diosque, P. Elucidating diversity in the class composition of the minicircle hypervariable region of Trypanosoma cruzi: New perspectives on typing and kDNA inheritance. PLoS Negl. Trop. Dis. 2019, 13, e0007536. [Google Scholar] [CrossRef]
- Balouz, V.; Bracco, L.; Ricci, A.D.; Romer, G.; Agüero, F.; Buscaglia, C.A. Serological Approaches for Trypanosoma cruzi Strain Typing. Trends Parasitol. 2021, 37, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Perez, A.; Poveda, C.; Ramírez, J.D.; Norman, F.; Gironés, N.; Guhl, F.; Monge-Maillo, B.; Fresno, M.; López-Vélez, R. Prevalence of Trypanosoma cruzi’s Discrete Typing Units in a cohort of Latin American migrants in Spain. Acta Trop. 2016, 157, 145–150. [Google Scholar] [CrossRef]
- Elias, M.C.; Vargas, N.S.; Zingales, B.; Schenkman, S. Organization of satellite DNA in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2003, 129, 1–9. [Google Scholar] [CrossRef]
- Elias, M.C.; Vargas, N.; Tomazi, L.; Pedroso, A.; Zingales, B.; Schenkman, S.; Briones, M.R. Comparative analysis of genomic sequences suggests that Trypanosoma cruzi CL Brener contains two sets of non-intercalated repeats of satellite DNA that correspond to T. cruzi I and T. cruzi II types. Mol. Biochem. Parasitol. 2005, 140, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Vago, A.R.; Andrade, L.O.; Leite, A.A.; d’Avila Reis, D.; Macedo, A.M.; Adad, S.J.; Tostes, S., Jr.; Moreira, M.C.; Filho, G.B.; Pena, S.D. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: Differential distribution of genetic types into diverse organs. Am. J. Pathol. 2000, 156, 1805–1809. [Google Scholar] [CrossRef] [PubMed]
- Macedo, A.M.; Pena, S.D. Genetic Variability of Trypanosoma cruzi: Implications for the Pathogenesis of Chagas Disease. Parasitol. Today 1998, 14, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Rojo, G.; Pèlissier, F.; Sandoval-Rodriguez, A.; Bacigalupo, A.; García, V.; Pinto, R.; Ortiz, S.; Botto-Mahan, C.; Cattan, P.E.; Solari, A. Organs infected with Trypanosoma cruzi and DTU identification in the naturally infected rodent Octodon degus. Exp. Parasitol. 2020, 215, 107931. [Google Scholar] [CrossRef]
- Flores-Villegas, A.L.; Jiménez-Cortés, J.G.; González, J.; Moreno-Rodríguez, A.; Pérez-Cabeza de Vaca, R.; Segal-Kischinevzky, C.; Bucio-Torres, M.I.; De Fuentes-Vicente, J.A.; Nava-Lazaro, E.; Salazar-Schettino, P.M.; et al. Parasitemia and Differential Tissue Tropism in Mice Infected with Trypanosoma cruzi Isolates Obtained from Meccus phyllosoma in the State of Oaxaca, Mexico. Pathogens 2022, 11, 1141. [Google Scholar] [CrossRef] [PubMed]
- D’Avila, D.A.; Macedo, A.M.; Valadares, H.M.; Gontijo, E.D.; de Castro, A.M.; Machado, C.R.; Chiari, E.; Galvão, L.M. Probing population dynamics of Trypanosoma cruzi during progression of the chronic phase in chagasic patients. J. Clin. Microbiol. 2009, 47, 1718–1725. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llovera, A.; Abras, A.; Fernández-Arévalo, A.; Ballart, C.; Heras, S.; Muñoz, C.; Gállego, M. Genetic Diversity of Trypanosoma cruzi in the United States of America: The Least Endemic Country for Chagas Disease. Life 2024, 14, 901. https://doi.org/10.3390/life14070901
Llovera A, Abras A, Fernández-Arévalo A, Ballart C, Heras S, Muñoz C, Gállego M. Genetic Diversity of Trypanosoma cruzi in the United States of America: The Least Endemic Country for Chagas Disease. Life. 2024; 14(7):901. https://doi.org/10.3390/life14070901
Chicago/Turabian StyleLlovera, Arnau, Alba Abras, Anna Fernández-Arévalo, Cristina Ballart, Sandra Heras, Carmen Muñoz, and Montserrat Gállego. 2024. "Genetic Diversity of Trypanosoma cruzi in the United States of America: The Least Endemic Country for Chagas Disease" Life 14, no. 7: 901. https://doi.org/10.3390/life14070901
APA StyleLlovera, A., Abras, A., Fernández-Arévalo, A., Ballart, C., Heras, S., Muñoz, C., & Gállego, M. (2024). Genetic Diversity of Trypanosoma cruzi in the United States of America: The Least Endemic Country for Chagas Disease. Life, 14(7), 901. https://doi.org/10.3390/life14070901






