Gut Microbiota in the Progression of Type 2 Diabetes and the Potential Role of Exercise: A Critical Review
Abstract
:1. Introduction
2. Pathophysiology of Type 2 Diabetes
3. Gut Microbiota and Type 2 Diabetes
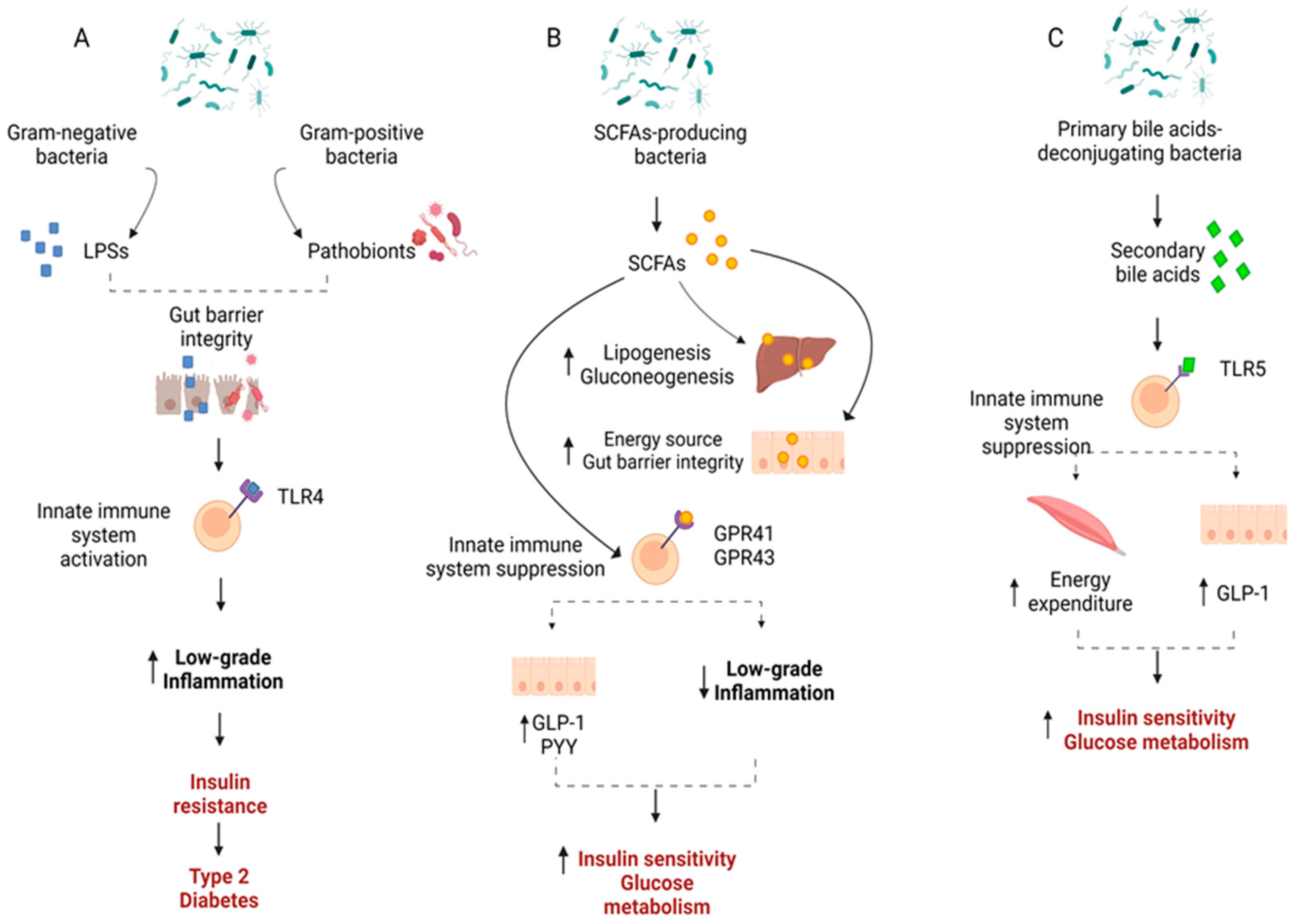
4. Mechanisms through Which Gut Microbiota May Affect Type 2 Diabetes
5. Gut Microbiota in the Progression from Normal, to Pre-Diabetes, to Type 2 Diabetes
6. The Potential Role of Exercise on Gut Microbiota in Pre-Diabetes and Type 2 Diabetes
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas—10th Edition. Available online: https://diabetesatlas.org/atlas/tenth-edition/ (accessed on 30 June 2024).
- World Health Organization; International Diabetes Federation. Definition and Diagnosis of Diabetes Mellitus and Intermediate Hyperglycaemia: Report of a WHO/IDF Consultation; WHO: Geneva, Switzerland, 2006. [Google Scholar]
- Stumvoll, M.; Goldstein, B.J.; van Haeften, T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet 2005, 365, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yang, Y.; Li, Y.; Han, R. Physical Exercise as Therapy for Type 2 Diabetes Mellitus: From Mechanism to Orientation. Ann. Nutr. Metab. 2019, 74, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, J.; Izquierdo, M.; Arguelles, I.; Forga, L.; Larrion, J.L.; Garcia-Unciti, M.; Idoate, F.; Gorostiaga, E.M. Twice-weekly progressive resistance training decreases abdominal fat and improves insulin sensitivity in older men with type 2 diabetes. Diabetes Care 2005, 28, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.I.; Goodyear, L.J. Exercise and type 2 diabetes: Molecular mechanisms regulating glucose uptake in skeletal muscle. Adv. Physiol. Educ. 2014, 38, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Winnick, J.J.; Sherman, W.M.; Habash, D.L.; Stout, M.B.; Failla, M.L.; Belury, M.A.; Schuster, D.P. Short-term aerobic exercise training in obese humans with type 2 diabetes mellitus improves whole-body insulin sensitivity through gains in peripheral, not hepatic insulin sensitivity. J. Clin. Endocrinol. Metab. 2008, 93, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.M.; Davy, B.M.; Hulver, M.W.; Neilson, A.P.; Bennett, B.J.; Davy, K.P. Does Exercise Alter Gut Microbial Composition? A Systematic Review. Med. Sci. Sports Exerc. 2019, 51, 160–167. [Google Scholar] [CrossRef]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 2014, 105, 141–150. [Google Scholar] [CrossRef]
- Guthrie, R.A.; Guthrie, D.W. Pathophysiology of diabetes mellitus. Crit. Care Nurs. Q. 2004, 27, 113–125. [Google Scholar] [CrossRef]
- DeFronzo, R.A.; Tripathy, D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care 2009, 32 (Suppl. 2), S157–S163. [Google Scholar] [CrossRef] [PubMed]
- Shulman, G.I.; Rothman, D.L.; Jue, T.; Stein, P.; DeFronzo, R.A.; Shulman, R.G. Quantitation of muscle glycogen synthesis in normal subjects and subjects with non-insulin-dependent diabetes by 13C nuclear magnetic resonance spectroscopy. N. Engl. J. Med. 1990, 322, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Shulman, G.I. Mechanisms of Insulin Action and Insulin Resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef] [PubMed]
- Faerch, K.; Hulman, A.; Solomon, T.P. Heterogeneity of Pre-diabetes and Type 2 Diabetes: Implications for Prediction, Prevention and Treatment Responsiveness. Curr. Diabetes Rev. 2016, 12, 30–41. [Google Scholar] [CrossRef] [PubMed]
- D‘Alessio, D. The role of dysregulated glucagon secretion in type 2 diabetes. Diabetes Obes. Metab. 2011, 13 (Suppl. S1), 126–132. [Google Scholar] [CrossRef]
- Haedersdal, S.; Lund, A.; Knop, F.K.; Vilsboll, T. The Role of Glucagon in the Pathophysiology and Treatment of Type 2 Diabetes. Mayo Clin. Proc. 2018, 93, 217–239. [Google Scholar] [CrossRef]
- Del Prato, S.; Castellino, P.; Simonson, D.C.; DeFronzo, R.A. Hyperglucagonemia and insulin-mediated glucose metabolism. J. Clin. Investig. 1987, 79, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Faerch, K.; Borch-Johnsen, K.; Holst, J.J.; Vaag, A. Pathophysiology and aetiology of impaired fasting glycaemia and impaired glucose tolerance: Does it matter for prevention and treatment of type 2 diabetes? Diabetologia 2009, 52, 1714–1723. [Google Scholar] [CrossRef]
- Faerch, K.; Vaag, A.; Holst, J.J.; Hansen, T.; Jørgensen, T.; Borch-Johnsen, K. Natural history of insulin sensitivity and insulin secretion in the progression from normal glucose tolerance to impaired fasting glycemia and impaired glucose tolerance: The Inter99 study. Diabetes Care 2009, 32, 439–444. [Google Scholar] [CrossRef]
- Larsson, H.; Ahren, B. Islet dysfunction in insulin resistance involves impaired insulin secretion and increased glucagon secretion in postmenopausal women with impaired glucose tolerance. Diabetes Care 2000, 23, 650–657. [Google Scholar] [CrossRef]
- Lund, A.; Bagger, J.I.; Christensen, M.; Knop, F.K.; Vilsboll, T. Glucagon and type 2 diabetes: The return of the alpha cell. Curr. Diabetes Rep. 2014, 14, 555. [Google Scholar] [CrossRef]
- Holst, J.J. The physiology of glucagon-like peptide 1. Physiol. Rev. 2007, 87, 1409–1439. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J., Jr.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef]
- Færch, K.; Torekov, S.S.; Vistisen, D.; Johansen, N.B.; Witte, D.R.; Jonsson, A.; Pedersen, O.; Hansen, T.; Lauritzen, T.; Sandbæk, A.; et al. GLP-1 Response to Oral Glucose Is Reduced in Prediabetes, Screen-Detected Type 2 Diabetes, and Obesity and Influenced by Sex: The ADDITION-PRO Study. Diabetes 2015, 64, 2513–2525. [Google Scholar] [CrossRef]
- Donath, M.Y.; Shoelson, S.E. Type 2 diabetes as an inflammatory disease. Nat. Rev. Immunol. 2011, 11, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Brannick, B.; Wynn, A.; Dagogo-Jack, S. Prediabetes as a toxic environment for the initiation of microvascular and macrovascular complications. Exp. Biol. Med. 2016, 241, 1323–1331. [Google Scholar] [CrossRef]
- Olson, N.C.; Callas, P.W.; Hanley, A.J.; Festa, A.; Haffner, S.M.; Wagenknecht, L.E.; Tracy, R.P. Circulating levels of TNF-α are associated with impaired glucose tolerance, increased insulin resistance, and ethnicity: The Insulin Resistance Atherosclerosis Study. J. Clin. Endocrinol. Metab. 2012, 97, 1032–1040. [Google Scholar] [CrossRef]
- Sabanayagam, C.; Shankar, A.; Lim, S.C.; Lee, J.; Tai, E.S.; Wong, T.Y. Serum C-reactive protein level and prediabetes in two Asian populations. Diabetologia 2011, 54, 767–775. [Google Scholar] [CrossRef]
- Chawla, A.; Nguyen, K.D.; Goh, Y.P. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 2011, 11, 738–749. [Google Scholar] [CrossRef]
- Spranger, J.; Kroke, A.; Mohlig, M.; Hoffmann, K.; Bergmann, M.M.; Ristow, M.; Boeing, H.; Pfeiffer, A.F. Inflammatory cytokines and the risk to develop type 2 diabetes: Results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes 2003, 52, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Shoelson, S.E.; Lee, J.; Goldfine, A.B. Inflammation and insulin resistance. J. Clin. Investig. 2006, 116, 1793–1801. [Google Scholar] [CrossRef]
- Luc, K.; Schramm-Luc, A.; Guzik, T.J.; Mikolajczyk, T.P. Oxidative stress and inflammatory markers in prediabetes and diabetes. J. Physiol. Pharmacol. 2019, 70, 809–824. [Google Scholar] [CrossRef]
- Bashan, N.; Kovsan, J.; Kachko, I.; Ovadia, H.; Rudich, A. Positive and negative regulation of insulin signaling by reactive oxygen and nitrogen species. Physiol. Rev. 2009, 89, 27–71. [Google Scholar] [CrossRef]
- Rains, J.L.; Jain, S.K. Oxidative stress, insulin signaling, and diabetes. Free Radic. Biol. Med. 2011, 50, 567–575. [Google Scholar] [CrossRef]
- Batrakoulis, A.; Jamurtas, A.Z.; Draganidis, D.; Georgakouli, K.; Tsimeas, P.; Poulios, A.; Syrou, N.; Deli, C.K.; Papanikolaou, K.; Tournis, S.; et al. Hybrid Neuromuscular Training Improves Cardiometabolic Health and Alters Redox Status in Inactive Overweight and Obese Women: A Randomized Controlled Trial. Antioxidants 2021, 10, 1601. [Google Scholar] [CrossRef]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Parte, A.C.; Sarda Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Goker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimaraes, V.; Sokol, H.; Dore, J.; Corthier, G.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- He, Y.; Wu, W.; Zheng, H.-M.; Li, P.; McDonald, D.; Sheng, H.-F.; Chen, M.-X.; Chen, Z.-H.; Ji, G.-Y.; Zheng, Z.-D.-X.; et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 2018, 24, 1532–1535. [Google Scholar] [CrossRef]
- Lang, J.M.; Eisen, J.A.; Zivkovic, A.M. The microbes we eat: Abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types. PeerJ 2014, 2, e659. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Wildeboer-Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67. [Google Scholar] [CrossRef]
- Raymond, F.; Ouameur, A.A.; Deraspe, M.; Iqbal, N.; Gingras, H.; Dridi, B.; Leprohon, P.; Plante, P.L.; Giroux, R.; Berube, E.; et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 2016, 10, 707–720. [Google Scholar] [CrossRef]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K.; et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Bressa, C.; Bailen-Andrino, M.; Perez-Santiago, J.; Gonzalez-Soltero, R.; Perez, M.; Montalvo-Lominchar, M.G.; Mate-Munoz, J.L.; Dominguez, R.; Moreno, D.; Larrosa, M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Xu, J.; Xu, C.; Chen, X.; Cai, X.; Yang, S.; Sheng, Y.; Wang, T. Regulation of an antioxidant blend on intestinal redox status and major microbiota in early weaned piglets. Nutrition 2014, 30, 584–589. [Google Scholar] [CrossRef]
- Aziz, Q.; Dore, J.; Emmanuel, A.; Guarner, F.; Quigley, E. Gut microbiota and gastrointestinal health: Current concepts and future directions. Neurogastroenterol. Motil. 2013, 25, 4–15. [Google Scholar] [CrossRef]
- Krajmalnik-Brown, R.; Ilhan, Z.E.; Kang, D.W.; DiBaise, J.K. Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. Off. Publ. Am. Soc. Parenter. Enter. Nutr. 2012, 27, 201–214. [Google Scholar] [CrossRef]
- Deli, C.K.; Poulios, A.; Georgakouli, K.; Papanikolaou, K.; Papoutsis, A.; Selemekou, M.; Karathanos, V.T.; Draganidis, D.; Tsiokanos, A.; Koutedakis, Y.; et al. The effect of pre-exercise ingestion of corinthian currant on endurance performance and blood redox status. J. Sports Sci. 2018, 36, 2172–2180. [Google Scholar] [CrossRef]
- Adeshirlarijaney, A.; Gewirtz, A.T. Considering gut microbiota in treatment of type 2 diabetes mellitus. Gut Microbes 2020, 11, 253–264. [Google Scholar] [CrossRef]
- Allin, K.H.; Nielsen, T.; Pedersen, O. Mechanisms in endocrinology: Gut microbiota in patients with type 2 diabetes mellitus. Eur. J. Endocrinol. 2015, 172, R167–R177. [Google Scholar] [CrossRef]
- Chen, J.; Thomsen, M.; Vitetta, L. Interaction of gut microbiota with dysregulation of bile acids in the pathogenesis of nonalcoholic fatty liver disease and potential therapeutic implications of probiotics. J. Cell Biochem. 2019, 120, 2713–2720. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, Y.; Xia, F.; Abudukerimu, B.; Zhang, W.; Guo, Y.; Wang, N.; Lu, Y. A Glucagon-Like Peptide-1 Receptor Agonist Lowers Weight by Modulating the Structure of Gut Microbiota. Front. Endocrinol. 2018, 9, 233. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, H.; Zhao, C.; Shi, Z.; Qiu, L.; Yang, F.; Zhou, X.; Han, X.; Wu, K.; Zhong, H.; et al. Metagenomic analysis reveals crosstalk between gut microbiota and glucose-lowering drugs targeting the gastrointestinal tract in Chinese patients with type 2 diabetes: A 6 month, two-arm randomised trial. Diabetologia 2022, 65, 1613–1626. [Google Scholar] [CrossRef]
- Chavez-Carbajal, A.; Pizano-Zarate, M.L.; Hernandez-Quiroz, F.; Ortiz-Luna, G.F.; Morales-Hernandez, R.M.; De Sales-Millan, A.; Hernandez-Trejo, M.; Garcia-Vite, A.; Beltran-Lagunes, L.; Hoyo-Vadillo, C.; et al. Characterization of the Gut Microbiota of Individuals at Different T2D Stages Reveals a Complex Relationship with the Host. Microorganisms 2020, 8, 94. [Google Scholar] [CrossRef]
- Egshatyan, L.; Kashtanova, D.; Popenko, A.; Tkacheva, O.; Tyakht, A.; Alexeev, D.; Karamnova, N.; Kostryukova, E.; Babenko, V.; Vakhitova, M.; et al. Gut microbiota and diet in patients with different glucose tolerance. Endocr. Connect. 2016, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gaike, A.H.; Paul, D.; Bhute, S.; Dhotre, D.P.; Pande, P.; Upadhyaya, S.; Reddy, Y.; Sampath, R.; Ghosh, D.; Chandraprabha, D.; et al. The Gut Microbial Diversity of Newly Diagnosed Diabetics but Not of Prediabetics Is Significantly Different from That of Healthy Nondiabetics. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Inoue, R.; Ohue-Kitano, R.; Tsukahara, T.; Tanaka, M.; Masuda, S.; Inoue, T.; Yamakage, H.; Kusakabe, T.; Hasegawa, K.; Shimatsu, A.; et al. Prediction of functional profiles of gut microbiota from 16S rRNA metagenomic data provides a more robust evaluation of gut dysbiosis occurring in Japanese type 2 diabetic patients. J. Clin. Biochem. Nutr. 2017, 61, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, S.M.; Carson, T.; Lowe, J.; Ramaraj, T.; Leff, J.W.; Luo, L.; Bell, C.J.; Shah, V.O. Composition, Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. J. Diabetes Obes. 2015, 2, 1–7. [Google Scholar] [CrossRef]
- Sedighi, M.; Razavi, S.; Navab-Moghadam, F.; Khamseh, M.E.; Alaei-Shahmiri, F.; Mehrtash, A.; Amirmozafari, N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017, 111, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lou, H.; Peng, Y.; Chen, S.; Zhang, Y.; Li, X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine 2019, 66, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Ren, H.; Lu, Y.; Fang, C.; Hou, G.; Yang, Z.; Chen, B.; Yang, F.; Zhao, Y.; Shi, Z.; et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. EBioMedicine 2019, 47, 373–383. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Mao, X.; Tao, Y.; Ran, X.; Zhao, H.; Xiong, J.; Li, L. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks. PLoS ONE 2017, 12, e0172774. [Google Scholar] [CrossRef]
- Gravdal, K.; Kirste, K.H.; Grzelak, K.; Kirubakaran, G.T.; Leissner, P.; Saliou, A.; Casèn, C. Exploring the gut microbiota in patients with pre-diabetes and treatment naïve diabetes type 2—A pilot study. BMC Endocr. Disord. 2023, 23, 179. [Google Scholar] [CrossRef]
- Zhang, Z.; Tian, T.; Chen, Z.; Liu, L.; Luo, T.; Dai, J. Characteristics of the gut microbiome in patients with prediabetes and type 2 diabetes. PeerJ 2021, 9, e10952. [Google Scholar] [CrossRef]
- Diener, C.; Reyes-Escogido, M.L.; Jimenez-Ceja, L.M.; Matus, M.; Gomez-Navarro, C.M.; Chu, N.D.; Zhong, V.; Tejero, M.E.; Alm, E.; Resendis-Antonio, O.; et al. Progressive Shifts in the Gut Microbiome Reflect Prediabetes and Diabetes Development in a Treatment-Naive Mexican Cohort. Front. Endocrinol. 2020, 11, 602326. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Tremaroli, V.; Schmidt, C.; Lundqvist, A.; Olsson, L.M.; Krämer, M.; Gummesson, A.; Perkins, R.; Bergström, G.; Bäckhed, F. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab. 2020, 32, 379–390.e373. [Google Scholar] [CrossRef]
- Zhang, S.; Cai, Y.; Meng, C.; Ding, X.; Huang, J.; Luo, X.; Cao, Y.; Gao, F.; Zou, M. The role of the microbiome in diabetes mellitus. Diabetes Res. Clin. Pract. 2021, 172, 108645. [Google Scholar] [CrossRef] [PubMed]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Kedenko, I.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ma, C.; Han, L.; Nawaz, M.; Gao, F.; Zhang, X.; Yu, P.; Zhao, C.; Li, L.; Zhou, A.; et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr. Microbiol. 2010, 61, 69–78. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Greer, R.L.; Dong, X.; Moraes, A.C.; Zielke, R.A.; Fernandes, G.R.; Peremyslova, E.; Vasquez-Perez, S.; Schoenborn, A.A.; Gomes, E.P.; Pereira, A.C.; et al. Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism. Nat. Commun. 2016, 7, 13329. [Google Scholar] [CrossRef]
- Zhang, L.; Qin, Q.; Liu, M.; Zhang, X.; He, F.; Wang, G. Akkermansia muciniphila can reduce the damage of gluco/lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog. Dis. 2018, 76, fty028. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Yassour, M.; Lim, M.Y.; Yun, H.S.; Tickle, T.L.; Sung, J.; Song, Y.M.; Lee, K.; Franzosa, E.A.; Morgan, X.C.; Gevers, D.; et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Tuovinen, E.; Keto, J.; Nikkila, J.; Matto, J.; Lahteenmaki, K. Cytokine response of human mononuclear cells induced by intestinal Clostridium species. Anaerobe 2013, 19, 70–76. [Google Scholar] [CrossRef]
- Cani, P.D.; Possemiers, S.; Van de Wiele, T.; Guiot, Y.; Everard, A.; Rottier, O.; Geurts, L.; Naslain, D.; Neyrinck, A.; Lambert, D.M.; et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009, 58, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Moya-Perez, A.; Neef, A.; Sanz, Y. Bifidobacterium pseudocatenulatum CECT 7765 Reduces Obesity-Associated Inflammation by Restoring the Lymphocyte-Macrophage Balance and Gut Microbiota Structure in High-Fat Diet-Fed Mice. PLoS ONE 2015, 10, e0126976. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Huh, C.S.; Choi, I.D.; Jeong, J.W.; Ku, H.K.; Ra, J.H.; Kim, T.Y.; Kim, G.B.; Sim, J.H.; Ahn, Y.T. The anti-diabetic activity of Bifidobacterium lactis HY8101 in vitro and in vivo. J. Appl. Microbiol. 2014, 117, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.W.; Pham, H.-P.; Bridonneau, C.; Aubry, C.; Lamas, B.; Martin-Gallausiaux, C.; Moroldo, M.; Rainteau, D.; Lapaque, N.; Six, A.; et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J. 2016, 10, 460–477. [Google Scholar] [CrossRef]
- Zhu, C.; Song, K.; Shen, Z.; Quan, Y.; Tan, B.; Luo, W.; Wu, S.; Tang, K.; Yang, Z.; Wang, X. Roseburia intestinalis inhibits interleukin17 excretion and promotes regulatory T cells differentiation in colitis. Mol. Med. Rep. 2018, 17, 7567–7574. [Google Scholar] [CrossRef]
- Tian, P.; Li, B.; He, C.; Song, W.; Hou, A.; Tian, S.; Meng, X.; Li, K.; Shan, Y. Antidiabetic (type 2) effects of Lactobacillus G15 and Q14 in rats through regulation of intestinal permeability and microbiota. Food Funct. 2016, 7, 3789–3797. [Google Scholar] [CrossRef]
- König, J.; Wells, J.; Cani, P.D.; García-Ródenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.J. Human Intestinal Barrier Function in Health and Disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef]
- Chelakkot, C.; Choi, Y.; Kim, D.K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.S.; Jee, Y.K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Emoto, T.; Yamashita, T.; Watanabe, H.; Hayashi, T.; Tabata, T.; Hoshi, N.; Hatano, N.; Ozawa, G.; Sasaki, N.; et al. Bacteroides vulgatus and Bacteroides dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation 2018, 138, 2486–2498. [Google Scholar] [CrossRef]
- Carlsson, A.H.; Yakymenko, O.; Olivier, I.; Håkansson, F.; Postma, E.; Keita, A.V.; Söderholm, J.D. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand. J. Gastroenterol. 2013, 48, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Aw, W.; Fukuda, S. Understanding the role of the gut ecosystem in diabetes mellitus. J. Diabetes Investig. 2018, 9, 5–12. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat. Metab. 2020, 2, 840–848. [Google Scholar] [CrossRef]
- Chambers, E.S.; Viardot, A.; Psichas, A.; Morrison, D.J.; Murphy, K.G.; Zac-Varghese, S.E.; MacDougall, K.; Preston, T.; Tedford, C.; Finlayson, G.S.; et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 2015, 64, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Burger-van Paassen, N.; Vincent, A.; Puiman, P.; van der Sluis, M.; Bouma, J.; Boehm, G.; van Goudoever, H.; Van Seuningen, I.; Renes, I.B. The Regulation of the Intestinal Mucin MUC2 Expression by Short Chain Fatty Acids: Implications for Epithelial Protection. Gastroenterology 2009, 136, A41. [Google Scholar]
- Kinoshita, M.; Suzuki, Y.; Saito, Y. Butyrate reduces colonic paracellular permeability by enhancing PPARgamma activation. Biochem. Biophys. Res. Commun. 2002, 293, 827–831. [Google Scholar] [CrossRef]
- Peng, L.; Li, Z.R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef]
- Segain, J.P.; Raingeard de la Blétière, D.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottière, H.M.; Galmiche, J.P. Butyrate inhibits inflammatory responses through NFkappaB inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Di, Y.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef]
- Vrieze, A.; Out, C.; Fuentes, S.; Jonker, L.; Reuling, I.; Kootte, R.S.; van Nood, E.; Holleman, F.; Knaapen, M.; Romijn, J.A.; et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 2014, 60, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Allin, K.H.; Tremaroli, V.; Caesar, R.; Jensen, B.A.H.; Damgaard, M.T.F.; Bahl, M.I.; Licht, T.R.; Hansen, T.H.; Nielsen, T.; Dantoft, T.M.; et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia 2018, 61, 810–820. [Google Scholar] [CrossRef]
- Nuli, R.; Cai, J.; Kadeer, A.; Zhang, Y.; Mohemaiti, P. Integrative Analysis Toward Different Glucose Tolerance-Related Gut Microbiota and Diet. Front. Endocrinol. 2019, 10, 295. [Google Scholar] [CrossRef]
- Ghaemi, F.; Fateh, A.; Sepahy, A.A.; Zangeneh, M.; Ghanei, M.; Siadat, S.D. Intestinal Microbiota Composition in Iranian Diabetic, Pre-diabetic and Healthy Individuals. J. Diabetes Metab. Disord. 2020, 19, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, X.; Xu, X.; Ming, J.; Wang, Z.; Gao, B.; Xing, Y.; Zhou, J.; Fu, J.; Liu, T.; et al. The Fecal Microbiota Is Already Altered in Normoglycemic Individuals Who Go on to Have Type 2 Diabetes. Front. Cell Infect. Microbiol. 2021, 11, 598672. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Colberg, S.R.; Sigal, R.J.; Fernhall, B.; Regensteiner, J.G.; Blissmer, B.J.; Rubin, R.R.; Chasan-Taber, L.; Albright, A.L.; Braun, B. Exercise and type 2 diabetes: The American College of Sports Medicine and the American Diabetes Association: Joint position statement. Diabetes Care 2010, 33, e147–e167. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, J.P.; Sacks, J.; Nieuwoudt, S. The essential role of exercise in the management of type 2 diabetes. Clevel. Clin. J. Med. 2017, 84, S15–S21. [Google Scholar] [CrossRef] [PubMed]
- Batrakoulis, A.; Jamurtas, A.Z.; Georgakouli, K.; Draganidis, D.; Deli, C.K.; Papanikolaou, K.; Avloniti, A.; Chatzinikolaou, A.; Leontsini, D.; Tsimeas, P.; et al. High intensity, circuit-type integrated neuromuscular training alters energy balance and reduces body mass and fat in obese women: A 10-month training-detraining randomized controlled trial. PLoS ONE 2018, 13, e0202390. [Google Scholar] [CrossRef]
- Church, T.S.; Blair, S.N.; Cocreham, S.; Johannsen, N.; Johnson, W.; Kramer, K.; Mikus, C.R.; Myers, V.; Nauta, M.; Rodarte, R.Q.; et al. Effects of aerobic and resistance training on hemoglobin A1c levels in patients with type 2 diabetes: A randomized controlled trial. JAMA 2010, 304, 2253–2262. [Google Scholar] [CrossRef] [PubMed]
- Ben Ounis, O.; Elloumi, M.; Lac, G.; Makni, E.; Van Praagh, E.; Zouhal, H.; Tabka, Z.; Amri, M. Two-month effects of individualized exercise training with or without caloric restriction on plasma adipocytokine levels in obese female adolescents. Ann. D‘Endocrinol. 2009, 70, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Tofas, T.; Fatouros, I.G.; Draganidis, D.; Deli, C.K.; Chatzinikolaou, A.; Tziortzis, C.; Panayiotou, G.; Koutedakis, Y.; Jamurtas, A.Z. Effects of Cardiovascular, Resistance and Combined Exercise Training on Cardiovascular, Performance and Blood Redox Parameters in Coronary Artery Disease Patients: An 8-Month Training-Detraining Randomized Intervention. Antioxidants 2021, 10, 409. [Google Scholar] [CrossRef]
- Silva, J.S.C.; Seguro, C.S.; Naves, M.M.V. Gut microbiota and physical exercise in obesity and diabetes—A systematic review. Nutr. Metab. Cardiovasc. Dis. 2022, 32, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.U.; Yassine, H.M.; Sohail, A.; Al Thani, A.A. Impact of Physical Exercise on Gut Microbiome, Inflammation, and the Pathobiology of Metabolic Disorders. Rev. Diabet. Stud. RDS 2019, 15, 35–48. [Google Scholar] [CrossRef]
- Estaki, M.; Pither, J.; Baumeister, P.; Little, J.P.; Gill, S.K.; Ghosh, S.; Ahmadi-Vand, Z.; Marsden, K.R.; Gibson, D.L. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 2016, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Lambert, J.E.; Myslicki, J.P.; Bomhof, M.R.; Belke, D.D.; Shearer, J.; Reimer, R.A. Exercise training modifies gut microbiota in normal and diabetic mice. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2015, 40, 749–752. [Google Scholar] [CrossRef]
- Pasini, E.; Corsetti, G.; Assanelli, D.; Testa, C.; Romano, C.; Dioguardi, F.S.; Aquilani, R. Effects of chronic exercise on gut microbiota and intestinal barrier in human with type 2 diabetes. Minerva Medica 2019, 110, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Motiani, K.K.; Collado, M.C.; Eskelinen, J.J.; Virtanen, K.A.; Loyttyniemi, E.; Salminen, S.; Nuutila, P.; Kalliokoski, K.K.; Hannukainen, J.C. Exercise Training Modulates Gut Microbiota Profile and Improves Endotoxemia. Med. Sci. Sports Exerc. 2020, 52, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Torquati, L.; Gajanand, T.; Cox, E.R.; Willis, C.R.G.; Zaugg, J.; Keating, S.E.; Coombes, J.S. Effects of exercise intensity on gut microbiome composition and function in people with type 2 diabetes. Eur. J. Sport Sci. 2023, 23, 530–541. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Ni, Y.; Cheung, C.K.Y.; Lam, K.S.L.; Wang, Y.; Xia, Z.; Ye, D.; Guo, J.; Tse, M.A.; et al. Gut Microbiome Fermentation Determines the Efficacy of Exercise for Diabetes Prevention. Cell Metab. 2020, 31, 77–91.e75. [Google Scholar] [CrossRef]
- Bohm, A.; Weigert, C.; Staiger, H.; Haring, H.U. Exercise and diabetes: Relevance and causes for response variability. Endocrine 2016, 51, 390–401. [Google Scholar] [CrossRef]

| Study | Gut Microbiota Composition | Gut Microbiota Function | Associations of GM with T2D |
|---|---|---|---|
| Karlsson 2013 [66] | Species model ↑ 4 Lactobacillus species ↓ 5 Clostridium species Metagenomic clusters (MGCs) model ↑ Eubacteriales, 2 Clostridium clostridioforme, Lactobacillus gasseri, Streptococcus mutans ↓ Roseburia, 2 unknown Clostridium, 5 Eubacteriales, Eubacterium eligens, Coriobacteriaceae, Bacteroides intestinalis | ↑ pathways for starch and glucose metabolism, fructose and mannose metabolism, ABC transporters for amino acids, ions and simple sugars, glycerolipid metabolism and fatty acids biosynthesis, cysteine and methionine metabolism ↓ pathways for flagellar assembly and riboflavin metabolism | Positive associations
Negative associations
|
| Larsen 2010 [67] | ↑ Betaproteobacteria, Bacilli ↓ Bacillotta, Clostridia | Positive associations
| |
| Qin 2012 [68] | ↑ Bacteroides caccae, Clostridium hathewayi, Clostridium ramosum, Clostridium symbiosum, Eggerthella lenta, Escherichia coli, Akkermansia muciniphila, Desulfovibrio sp. ↓ Eubacteriales sp. SS3/4, E. rectale, Faecalibacterium prausnitzii, Roseburia intestinalis, Roseburia inulinivorans, Haemophilus parainfluenzae | ↑ pathways for membrane transport of sugars, branched-chain amino acid transport, methane metabolism, xenobiotics degradation and metabolism, sulfate reduction, oxidative stress resistance, drug resistance ↓ pathways for bacterial chemotaxis, flagellar assembly, butyrate biosynthesis, metabolism of cofactors and vitamins | |
| Chavez-Carbajal 2020 [72] | ↓ Richness, ≠ β-diversity ↑ Bacteroidota, Sutterella ↓ Bacillotta Community drivers Sutterella, Prevotella, Ruminococcus | Positive associations
Negative associations
| |
| Egshatyan 2016 [73] | ↑ Blautia, Serratia ↓ Verrucomicrobiota | Positive associations
Negative associations
| |
| Gaike 2020 [74] | ↓ α-diversity, ≠ β-diversity ↑ Bacillotta/Bacteroidota, Bacillotta, Pseudomonadota, Lactobacillus ↓ Bacteroidota, Verrucomicrobiota, Akkermansia, Blautia, Prevotella, Ruminococcus Community drivers Sutterella, Prevotella, Ruminococcus | Positive associations
Negative associations
| |
| Inoue 2017 [75] | ↓ Blautia | ↑ Pathways for glycolysis/gluconeogenesis, tyrosine metabolism, naphthalene degradation, insulin signaling, phenylalanine metabolism, butirosin and neomycin biosynthesis, drug metabolism-cytochrome P450, metabolism of xenobiotics by cytochrome P450, retinol metabolism, ethylbenzene degradation, proximal tubule bicarbonate reclamation ↓ pathway for oxidative phosphorylation | Negative associations
|
| Lambeth 2015 [76] | ↑ Collinsella, Enterobacteriaceae unknown genus | ||
| Sedighi 2017 [77] | ↑ Lactobacillus, Fusobacterium ↓ Bifidobacterium | ||
| Zhang 2013 [78] | ↓ α-diversity ↑ Bacillotta, Betaproteobacteria, Clostridia, Lachnospiraceae, Ruminococcus, Dorea, Prevotella, Collinsella, Subdoligranulum, Eubacterium, Sporobacter, Abiotrophia, Peptostreptococcus, Clostridialles, Clostridialles sp. SS3/4 ↓ Bacteroides, Streptococcus, F. prausnitzii, Haemophilus parainfluenzae, Roseburia, Megamonas | Positive associations
Negative associations
| |
| Zhao 2019 [79] | ↑ Bacillotta, Pseudomonadota, Eubacterium hallii, Eubacterium ventriosum, Ruminococcus torques, Blautia, Coprococcus, Lachnospiraceae, Sabdoligranulum, Dialister ↓ Bacteroidota, Bacteroides, Prevotella, Ruminococcus 2 Fecal Metabolites ↓ Isobutyrate, hexanoic acid, cholic acid, oleanolic acid ↑ Linolenic acid, diacylglycerol | ↓ pathways for tricarboxylic acid cycle, sugar metabolism ↑ pathways for glycerophospholipid metabolism, synthesis and degradation of ketone bodies, fatty acid metabolism | Positive associations
Negative associations
|
| Zong 2019 [80] | ↑ B. caccae, Bacteroides finegoldii, Collinsella intestinalis, Megasphaera elsdenii ↓ Dialister invisus, Roseburia hominis, E. coli, A. muciniphila, Clostridium bartlettii | ↑ Sugar phosphotransferase systems, ATP-binding cassette transporters of amino acids, bacterial secretion system | |
| Wang 2017 [81] | ↑Veillonellaceae ↓ Erysipelotrichaceae | ||
| Gravdal 2023 [82] | ↑ Dorea, Dorea formicigenerans, Dorea longicatena, Erysipelotrichia, Erysipelotrichales, Erysipelotrichaceae, Turicibacter, Turicibacter sanguinis ↓ Anaerotignum | ||
| Zhang 2021 [83] | ↑ Negativicutes, Finegoldia, Megasphaera, Prevotella, Alloprevotella ↓ Bacteroides, Paraprevotella, Helicobacter | ||
| Diener 2020 [84] | ↑ Escherichia/Shigella, Veillonella ↓ Anaerostipes, Blautia | Positive associations
Negative associations
|
| Study | From NGT to Pre-D | From Pre-D to T2D |
|---|---|---|
| Chavez-Carbajal 2020 [72] | ↓ Richness ↑Sutterella ↓ β-diversity | |
| Gaike 2020 [74] | Community drivers Blautia, Sutterella | ↑ Bacillotta/Bacteroidota |
| Inoue 2017 [75] | ↑ Blautia, Serratia ↑ Prevotella ↓ Verrucomicrobiota | ↑ Blautia, Serratia |
| Lambeth 2015 [76] | ↑ Chloracidobacteria, Pseudonocardiaceae unknown genus | ↑ Collinsella, unknown genus of Enterobacteriaceae ↓ Chloracidobacteria, Pseudonocardiaceae unknown genus |
| Zhang 2013 [78] | ↑ Betaproteobacteria, Prevotella, Megamonas, Eubacteriales sp. SS3/4 ↓ Verrucomicrobiota, Verrucomicrobiia, Roseburia, A. muciniphila, Streptococcus | ↑ Betaproteobacteria, Sutterella, Collinsella, Clostridia, Subdoligranulum, Clostridialles sp. SS3/4 ↓ Bacteroides, Streptococcus, Roseburia |
| Zong 2019 [80] | ↑ E. coli, Streptococcus salivarius, Eggerthella spp., Megasphaera elsdenii ↓ Dialister invisus, R. hominis, F. prausnitzii Functional pathways ↑ Sugar phosphotransferase systems, ATP- binding cassette transporters of amino acids, bacterial secretion system, microcin C transport system, autoinducer-2 (AI-2) transport system ↓ Modules of V-type ATPase, pyruvate:ferredoxin oxidoreductase, bacterial ribosomal proteins | ↑ F. prausnitzii, Bacteroides caccae, Bacteroides finegoldii, Collinsella intestinalis ↓ E. coli, A. muciniphila, C. bartlettii Functional pathways ↑ Modules of V-type ATPase, pyruvate:ferredoxin oxidoreductase, bacterial ribosomal proteins ↓ II–IV secretion system, AI-2 transport system |
| Gravdal 2023 [82] | ↑ Dorea spp., Pseudomonadota, Enterobacterales, Shigella spp., Escherichia spp. ↓ Roseburia spp., Roseburia intestinalis, Faecalibacterium prausnitzii, Dialister invisus, Veillonella spp., Sutterella wadsworthensis | N/A |
| Zhang 2021 [83] | ↑ Pseudomonadota, Haemophilus, Escherichia/Shigella, Megasphaera ↓ Lachnospira, Paraprevotella, Helicobacter | ↑ Negativicutes ↓ Finegoldia, Bacteroides |
| Diener 2020 [84] | ↑ Escherichia, Veillonella ↓ Anaerostipes, Anaerostipes hardus, Blautia | ↑ Escherichia, Veillonella ↓ Anaerostipes, Blautia |
| Allin 2018 [121] | ↑ Dorea, [Ruminococcus], Sutterella, Streptococcus ↓ Clostridium, Akkermansia muciniphila | N/A |
| Nuli 2019 [122] | ↓ α-diversity ↑Bacillotta, Actinomycetota, Saccharibacteria, Megamonas, Haemophilus, norank_p_Saccharibacteria ↓ Bacteroidota, Pseudomonadota, Ruminococcaceae, Barnesiella, Sutterella, Ruminiclostridium, Eubacteriales, Coriobacteriaceae, Flavonifractor | ↑ Deferribacteres, Moryella, Lachnospiraceae_NC2004_group |
| Ghaemi 2020 [123] | ↑ E. coli, Bacteroides fragilis ↓ F. prausnitzii | ↓ Bifidobacterium |
| Study | Study Group | Exercise Intervention | Changes in T2D Related Indices | Changes in GM Composition | Changes in GM Function |
|---|---|---|---|---|---|
| Lambert 2015 [136] | 6-week-old mice: T2D db/db sedentary, db/db exercise; Non-D: db/+ sedentary, db/+ exercise | 6 weeks of: (a) Moderate to high intensity treadmill running, 60–66 min/session, 5 days/week, or, (b) sedentary behavior | db/+ mice: ↑ Bifidobacterium spp., Lactobacillus spp., and Clostridium leptum, Clostridium cluster I; ↓ Bacteroides/Prevotella spp., and Methanobrevibacter spp. db/db mice: ↓ Total bacteria and Enterobacteriaceae, Bifidobacterium spp., Bacteroides/Prevotella spp., and Methanobrevibacter spp., Clostridium cluster I; ↑ Lactobacillus spp., and C. leptum | ||
| Pasini 2019 [137] | 30 male patients, with T2D for at least 2 years Medication Glucose, lipid, and blood pressure lowering agents | 6 months of: Aerobic, resistance and flexibility training, 90 min/session, 3 times/week | ↓ BW, BMI, fat mass, FPG, HOMA index, CRP, CHOL-T ↑ Lean body mass, VO2max | ↓ Candida albicans, and Mycetes spp. Intestinal barrier health ↓ Zonulin in feces | |
| Motiani 2020 [138] | 26 sedentary insulin-resistant male/ female patients with pre-D and T2D Medication No medication (n = 4), Metformin (n = 11), DPP-IV (n = 5), sulfonylurea (n = 1) | 2 weeks of: (a) Sprint interval training (SIT), 3 times/week, or, (b) Moderate intensity continuous training (MICT), 3 times/week | T2D related and functional indices SIT: ↑ VO2peak SIT & MICT: ↓ % BF, abdominal visceral fat mass, HbA1c, TNF-α, and LBP Intestinal fasting fatty acid uptake (IFAU) & glucose uptake (GU) SIT: ↔ GU, IFAU MICT: ↔ IGU, FAU in the duodenum or colon; ↓ FAU in the jejunum | SIT & MICT: ↓ Bacillotta/Bacteroidota ratio, Blautia spp., Clostridium spp.; ↑ Bacteroidota SIT: ↑ Lachnospira, Bacteroides MICT: ↑ Veillonella, Veillonella dispar, Faecalibacterium prausnitzii | |
| Torquati 2022 [139] | 12 T2D patients participating in <50 min/week of moderate intensity or 75 min/week of vigorous intensity physical activity Medication Metformin Or Metformin + DPP-4 inhibitors | 8 weeks of: (a) High intensity combined aerobic and resistance training, 26 min/session, 3 sessions/week (C-HIIT), or (b) Moderate intensity combined aerobic and resistance training, 52 min/session, 4 sessions/week (C-MICT) | C-HIIT vs. C-MICT: ↔FPG, HbA1c, CHO-T, HDL, LDL, TG, hs-CRP, SBP, DBP, BW, WC, VO2peak | C-MICT: ↑ Bifidobacterium, C. leptum, F. prausnitzii, A. municiphila, Escherichia spp., Enterococcus spp., Lachnospira eligens, Agathobaculum spp., Massimaliae spp. C-HIIT: ↑ Erysipelotrichales, Methanobrevibacter smithii, Oscillospirales, Ruminococcus bromii, Negativibacillus spp. | C-MICT: ↑ pathways for pyruvate metabolism (ko00620), AAs metabolism (ko00250, ko00260, ko00290) C-HIIT: ↓ pathways for bacterial secretion system—translocase/protein export SecD (K03072), Transcription regulation (Prokaryotic defense system K07316, Transcription factor K05499), Peptidase and inhibitors (K20608) |
| Liu 2020 [140] | 34 sedentary male individuals with pre-D (IGT and/or IFG) Medication: No antidiabetic medication, no antibiotics, no probiotics | 12 weeks of: (a) High intensity combined aerobic and strength training, 70 min/session, 3 sessions/week, or, (b) Sedentary behavior Responders and non-responders to exercise | Exercise group (whole group): ↓ Fasting INS, HOMA-IR, TG, CHO-T, resting HR, hs-CRP, BW, % BF Responders: ↓ 42.70% in fasting INS, 49.60% in HOMA-IR; ↑ 116.29% in Matsuda index Non-responders: ↔ | Responders: ↑ Lachnospiraceae, Streptococcus mitis; ↓ Akkermansia muciniphila, Alistipes putredinis, Alistipes shahii, Bacteroides cellulosilyticus, Bacteroides clarus, Bacteroides xylanisolvens Non-responders: ↑ A. shahii ↓ Escherichia coli, Ruminococcus gnavus | Responders: ↑pathways for quorum sensing, DNA replication, amino acid metabolism, glycan biosynthesis, lipid metabolism, BCAAs catabolism, SCFAs biosynthesis Non-responders: ↑ pathways for catabolism of AAAs and SAAs, production of colonic gases, and detrimental substrates for oxidative stress and inflammation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deli, C.K.; Fatouros, I.G.; Poulios, A.; Liakou, C.A.; Draganidis, D.; Papanikolaou, K.; Rosvoglou, A.; Gatsas, A.; Georgakouli, K.; Tsimeas, P.; et al. Gut Microbiota in the Progression of Type 2 Diabetes and the Potential Role of Exercise: A Critical Review. Life 2024, 14, 1016. https://doi.org/10.3390/life14081016
Deli CK, Fatouros IG, Poulios A, Liakou CA, Draganidis D, Papanikolaou K, Rosvoglou A, Gatsas A, Georgakouli K, Tsimeas P, et al. Gut Microbiota in the Progression of Type 2 Diabetes and the Potential Role of Exercise: A Critical Review. Life. 2024; 14(8):1016. https://doi.org/10.3390/life14081016
Chicago/Turabian StyleDeli, Chariklia K., Ioannis G. Fatouros, Athanasios Poulios, Christina A. Liakou, Dimitrios Draganidis, Konstantinos Papanikolaou, Anastasia Rosvoglou, Athanasios Gatsas, Kalliopi Georgakouli, Panagiotis Tsimeas, and et al. 2024. "Gut Microbiota in the Progression of Type 2 Diabetes and the Potential Role of Exercise: A Critical Review" Life 14, no. 8: 1016. https://doi.org/10.3390/life14081016







