The Domestication of Wild Boar Could Result in a Relaxed Selection for Maintaining Olfactory Capacity
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Collection
2.2. Transcriptional Profiling of Olfactory Mucosa
2.2.1. RNA Isolation
2.2.2. Data Processing and Differential Expression Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larson, G.; Fuller, D.Q. The Evolution of Animal Domestication. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 115–136. [Google Scholar] [CrossRef]
- Price, E.O. Behavioral Aspects of Animal Domestication. Q. Rev. Biol. 1984, 59, 1–32. [Google Scholar] [CrossRef]
- Lyons, S.; Amatangelo, K.L.; Behrensmeyer, A.K.; Bercovici, A.; Blois, J.L.; Davis, M.; DiMichele, W.A.; Du, A.; Eronen, J.T.; Tyler Faith, J.; et al. Holocene Shifts in the Assembly of Plant and Animal Communities Implicate Human Impacts. Nature 2016, 529, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Price, M.; Hongo, H. The Archaeology of Pig Domestication in Eurasia. J. Archaeol. Res. 2020, 28, 557–615. [Google Scholar] [CrossRef]
- Lega, C.; Raia, P.; Rook, L.; Fulgione, D. Size Matters: A Comparative Analysis of Pig Domestication. Holocene 2016, 26, 327–332. [Google Scholar] [CrossRef]
- Trut, L.; Oskina, I.; Kharlamova, A. Animal Evolution during Domestication: The Domesticated Fox as a Model. BioEssays 2009, 31, 349–360. [Google Scholar] [CrossRef]
- Van Laere, A.-S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A Regulatory Mutation in IGF2 Causes a Major QTL Effect on Muscle Growth in the Pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef]
- Fang, M.; Larson, G.; Soares Ribeiro, H.; Li, N.; Andersson, L. Contrasting Mode of Evolution at a Coat Color Locus in Wild and Domestic Pigs. PLoS Genet. 2009, 5, e1000341. [Google Scholar] [CrossRef]
- Zeder, M.A. Domestication as a Model System for the Extended Evolutionary Synthesis. Interface Focus 2017, 7, 20160133. [Google Scholar] [CrossRef]
- Albiach-Serrano, A.; Bräuer, J.; Cacchione, T.; Zickert, N.; Amici, F. The Effect of Domestication and Ontogeny in Swine Cognition (Sus Scrofa Scrofa and S. s. Domestica). Appl. Anim. Behav. Sci. 2012, 141, 25–35. [Google Scholar] [CrossRef]
- Kittawornrat, A.; Zimmerman, J.J. Toward a Better Understanding of Pig Behavior and Pig Welfare. Anim. Health. Res. Rev. 2011, 12, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, H.H.; Jones, R.B.; Schofield, C.P.; White, R.P.; Wathes, C.M. The Use of Olfactory and Other Cues for Social Recognition by Juvenile Pigs. Appl. Anim. Behav. Sci. 2001, 72, 321–333. [Google Scholar] [CrossRef]
- Brunjes, P.C.; Feldman, S.; Osterberg, S.K. The Pig Olfactory Brain: A Primer. Chem. Senses 2016, 41, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.-Q.; Webber, C.; Hehir-Kwa, J.; Pfundt, R.; Veltman, J.; Ponting, C.P. Reduced Purifying Selection Prevails over Positive Selection in Human Copy Number Variant Evolution. Genome Res. 2008, 18, 1711–1723. [Google Scholar] [CrossRef]
- Guerrier, S.; Coutinho-Budd, J.; Sassa, T.; Gresset, A.; Jordan, N.V.; Chen, K.; Jin, W.-L.; Frost, A.; Polleux, F. The F-BAR Domain of srGAP2 Induces Membrane Protrusions Required for Neuronal Migration and Morphogenesis. Cell 2009, 138, 990–1004. [Google Scholar] [CrossRef]
- Groenen, M.A.M.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.-J.; et al. Analyses of Pig Genomes Provide Insight into Porcine Demography and Evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Paudel, Y.; Madsen, O.; Megens, H.-J.; Frantz, L.A.F.; Bosse, M.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Copy Number Variation in the Speciation of Pigs: A Possible Prominent Role for Olfactory Receptors. BMC Genom. 2015, 16, 330. [Google Scholar] [CrossRef]
- Rinaldi, A. The Scent of Life: The Exquisite Complexity of the Sense of Smell in Animals and Humans. EMBO Rep. 2007, 8, 629–633. [Google Scholar] [CrossRef]
- Rekwot, P.I.; Ogwu, D.; Oyedipe, E.O.; Sekoni, V.O. The Role of Pheromones and Biostimulation in Animal Reproduction. Anim. Reprod. Sci. 2001, 65, 157–170. [Google Scholar] [CrossRef]
- Distel, H.; Hudson, R. The Contribution of the Olfactory and Tactile Modalities to the Nipple-Search Behaviour of Newborn Rabbits. J. Comp. Physiol. 1985, 157, 599–605. [Google Scholar] [CrossRef]
- Erdtmann, D.; Keuling, O. Behavioural Patterns of Free Roaming Wild Boar in a Spatiotemporal Context. PeerJ 2020, 8, e10409. [Google Scholar] [CrossRef] [PubMed]
- Fulgione, D.; Rippa, D.; Buglione, M.; Trapanese, M.; Petrelli, S.; Maselli, V. Unexpected but Welcome. Artificially Selected Traits May Increase Fitness in Wild Boar. Evol. Appl. 2016, 9, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Fulgione, D.; Buglione, M. The Boar War: Five Hot Factors Unleashing Boar Expansion and Related Emergency. Land 2022, 11, 887. [Google Scholar] [CrossRef]
- Wright, D. The Genetic Architecture of Domestication in Animals. Bioinform. Biol. Insights 2015, 9, 11–20. [Google Scholar] [CrossRef]
- Price, E.O. Behavioral Development in Animals Undergoing Domestication. Appl. Anim. Behav. Sci. 1999, 65, 245–271. [Google Scholar] [CrossRef]
- Diamond, J. Evolution, Consequences and Future of Plant and Animal Domestication. Nature 2002, 418, 700–707. [Google Scholar] [CrossRef]
- Albert, F.W.; Somel, M.; Carneiro, M.; Aximu-Petri, A.; Halbwax, M.; Thalmann, O.; Blanco-Aguiar, J.A.; Plyusnina, I.Z.; Trut, L.; Villafuerte, R.; et al. A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals. PLoS Genet. 2012, 8, e1002962. [Google Scholar] [CrossRef]
- Maselli, V.; Polese, G.; Larson, G.; Raia, P.; Forte, N.; Rippa, D.; Ligrone, R.; Vicidomini, R.; Fulgione, D. A Dysfunctional Sense of Smell: The Irreversibility of Olfactory Evolution in Free-Living Pigs. Evol. Biol. 2014, 41, 229–239. [Google Scholar] [CrossRef]
- Petrelli, S.; Buglione, M.; Maselli, V.; Troiano, C.; Larson, G.; Frantz, L.; Manin, A.; Ricca, E.; Baccigalupi, L.; Wright, D.; et al. Population Genomic, Olfactory, Dietary, and Gut Microbiota Analyses Demonstrate the Unique Evolutionary Trajectory of Feral Pigs. Mol. Ecol. 2021, 31, 220–237. [Google Scholar] [CrossRef]
- Petrelli, S.; Buglione, M.; Rivieccio, E.; Ricca, E.; Baccigalupi, L.; Scala, G.; Fulgione, D. Reprogramming of the Gut Microbiota Following Feralization in Sus Scrofa. Anim. Microbiome 2023, 5, 14. [Google Scholar] [CrossRef]
- Jacela, J.Y.; DeRouchey, J.M.; Tokach, M.D.; Goodband, R.D.; Nelssen, J.L.; Renter, D.G.; Dritz, S.S. Feed Additives for Swine: Fact Sheets—Flavors and Mold Inhibitors, Mycotoxin Binders, and Antioxidants (2010). Kans. Agric. Exp. Stn. Res. Rep. 2010, 27–32. [Google Scholar] [CrossRef]
- Peden, R.S.E.; Turner, S.P.; Boyle, L.A.; Camerlink, I. The Translation of Animal Welfare Research into Practice: The Case of Mixing Aggression between Pigs. Appl. Anim. Behav. Sci. 2018, 204, 1–9. [Google Scholar] [CrossRef]
- Schild, S.-L.A.; Rørvang, M.V. Pig Olfaction: The Potential Impact and Use of Odors in Commercial Pig Husbandry. Front. Anim. Sci. 2023, 4, 1215206. [Google Scholar] [CrossRef]
- Fraser, H.B. Genome-Wide Approaches to the Study of Adaptive Gene Expression Evolution: Systematic Studies of Evolutionary Adaptations Involving Gene Expression Will Allow Many Fundamental Questions in Evolutionary Biology to Be Addressed. BioEssays 2011, 33, 469–477. [Google Scholar] [CrossRef]
- McGregor, A.P.; Orgogozo, V.; Delon, I.; Zanet, J.; Srinivasan, D.G.; Payre, F.; Stern, D.L. Morphological Evolution through Multiple Cis-Regulatory Mutations at a Single Gene. Nature 2007, 448, 587–590. [Google Scholar] [CrossRef]
- Wray, G.A. The Evolutionary Significance of Cis-Regulatory Mutations. Nat. Rev. Genet. 2007, 8, 206–216. [Google Scholar] [CrossRef]
- Harrison, P.W.; Wright, A.E.; Mank, J.E. The Evolution of Gene Expression and the Transcriptome–Phenotype Relationship. Semin. Cell Dev. Biol. 2012, 23, 222–229. [Google Scholar] [CrossRef]
- Bosse, M.; Madsen, O.; Megens, H.-J.; Frantz, L.A.F.; Paudel, Y.; Crooijmans, R.P.M.A.; Groenen, M.A.M. Hybrid Origin of European Commercial Pigs Examined by an In-Depth Haplotype Analysis on Chromosome 1. Front. Genet. 2015, 5, 442. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maselli, V.; Rippa, D.; Deluca, A.; Larson, G.; Wilkens, B.; Linderholm, A.; Masseti, M.; Fulgione, D. Southern Italian Wild Boar Population, Hotspot of Genetic Diversity. Hystrix Ital. J. Mammal. 2016, 27, 137–144. [Google Scholar] [CrossRef]
- Keiter, D.A.; Mayer, J.J.; Beasley, J.C. What is in a “common” name? A call for consistent terminology for nonnative Sus scrofa. Wildl. Soc. Bull. 2016, 40, 384–387. [Google Scholar] [CrossRef]
- Albarella, U.; Tagliacozzo, A.; Dobney, K.; Rowley-Conwy, P. Pig Hunting and Husbandry in Prehistoric Italy: A Contribution to the Domestication Debate. Proc. Prehist. Soc. 2006, 72, 193–227. [Google Scholar] [CrossRef]
- Albarella, U.; Manconi, F.; Rowley-Conwy, P.; Vigne, J.D. Vigne Pigs of Corsica and Sardinia: A Biometrical Re-Evaluation of Their Status and History. In Archaeozoological Studies in Honour of Alfredo Riedel; Ufficio Beni Archeol: Bolzano, Italy, 2006; pp. 285–302. [Google Scholar]
- Albarella, U.; Dobney, K.; Ervynck, A. Pigs and Humans: 10,000 Years of Interaction; Oxford University Press: New York, NY, USA, 2007; ISBN 978-0-19-920704-6. [Google Scholar]
- Wilkens, B. Archeozoologia. Il Mediterraneo, La Storia, La Sardegna; Democratica Sarda: Sassari, Italy, 2012; ISBN 978-88-6025-251-7. [Google Scholar]
- Vigne. Les Mammifères Post-Glaciaires de Corse: Étude Archéozoologique; Supplément à “Gallia préhistoire”; Centre National de la Recherche Scientifique: Paris, France, 1989; ISBN 978-2-222-04130-6. [Google Scholar]
- Larson, G.; Dobney, K.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide Phylogeography of Wild Boar Reveals Multiple Centers of Pig Domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.C.; Wehr, N.H.; Litton, C.M. Wild Pigs in the Pacific Islands. In Invasive Wild Pigs in North America; VerCauteren, K.C., Beasley, J.C., Ditchkoff, S.S., Mayer, J.J., Roloff, G.J., Strickland, B.K., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 403–421. ISBN 978-1-315-23305-5. [Google Scholar]
- Maselli, V.; Rippa, D.; Russo, G.; Ligrone, R.; Soppelsa, O.; D’Aniello, B.; Raia, P.; Fulgione, D. Wild Boars’ Social Structure in the Mediterranean Habitat. Ital. J. Zool. 2014, 81, 610–617. [Google Scholar] [CrossRef]
- Frantz, L.A.F.; Schraiber, J.G.; Madsen, O.; Megens, H.-J.; Cagan, A.; Bosse, M.; Paudel, Y.; Crooijmans, R.P.M.A.; Larson, G.; Groenen, M.A.M. Evidence of Long-Term Gene Flow and Selection during Domestication from Analyses of Eurasian Wild and Domestic Pig Genomes. Nat. Genet. 2015, 47, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Gering, E.; Incorvaia, D.; Henriksen, R.; Conner, J.; Getty, T.; Wright, D. Getting Back to Nature: Feralization in Animals and Plants. Trends Ecol. Evol. 2019, 34, 1137–1151. [Google Scholar] [CrossRef] [PubMed]
- Iacolina, L.; Corlatti, L.; Buzan, E.; Safner, T.; Šprem, N. Hybridisation in European Ungulates: An Overview of the Current Status, Causes, and Consequences. Mammal Rev. 2019, 49, 45–59. [Google Scholar] [CrossRef]
- Matschke, G.H. Aging European Wild Hogs by Dentition. J. Wildl. Manag. 1967, 31, 109–113. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Evin, A.; Dobney, K.; Schafberg, R.; Owen, J.; Vidarsdottir, U.S.; Larson, G.; Cucchi, T. Phenotype and Animal Domestication: A Study of Dental Variation between Domestic, Wild, Captive, Hybrid and Insular Sus Scrofa. BMC Evol. Biol. 2015, 15, 6. [Google Scholar] [CrossRef]
- Frantz, L.A.F.; Haile, J.; Lin, A.T.; Scheu, A.; Geörg, C.; Benecke, N.; Alexander, M.; Linderholm, A.; Mullin, V.E.; Daly, K.G.; et al. Ancient Pigs Reveal a Near-Complete Genomic Turnover Following Their Introduction to Europe. Proc. Natl. Acad. Sci. USA 2019, 116, 17231–17238. [Google Scholar] [CrossRef]
- Masseti, M. The Economic Role of Sus in Early Human Fishing Communities. In Pigs and Humans; Oxford University Press: Oxford, UK, 2007; ISBN 978-0-19-920704-6. [Google Scholar]
- Gould, S.J. The Exaptive Excellence of Spandrels as a Term and Prototype. Proc. Natl. Acad. Sci. USA 1997, 94, 10750–10755. [Google Scholar] [CrossRef]
- Fulgione, D.; Trapanese, M.; Buglione, M.; Rippa, D.; Polese, G.; Maresca, V.; Maselli, V. Pre-Birth Sense of Smell in the Wild Boar: The Ontogeny of the Olfactory Mucosa. Zoology 2017, 123, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Zeder, M.A. Core Questions in Domestication Research. Proc. Natl. Acad. Sci. USA 2015, 112, 3191–3198. [Google Scholar] [CrossRef]
- Roncaglia, P.; Van Dam, T.J.P.; Christie, K.R.; Nacheva, L.; Toedt, G.; Huynen, M.A.; Huntley, R.P.; Gibson, T.J.; Lomax, J. The Gene Ontology of Eukaryotic Cilia and Flagella. Cilia 2017, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Sironen, A.; Shoemark, A.; Patel, M.; Loebinger, M.R.; Mitchison, H.M. Sperm Defects in Primary Ciliary Dyskinesia and Related Causes of Male Infertility. Cell. Mol. Life Sci. 2020, 77, 2029–2048. [Google Scholar] [CrossRef]
- Lehti, M.S.; Sironen, A. Formation and Function of Sperm Tail Structures in Association with Sperm Motility Defects. Biol. Reprod. 2017, 97, 522–536. [Google Scholar] [CrossRef]
- Leung, M.R.; Roelofs, M.C.; Ravi, R.T.; Maitan, P.; Henning, H.; Zhang, M.; Bromfield, E.G.; Howes, S.C.; Gadella, B.M.; Bloomfield-Gadêlha, H.; et al. The Multi-Scale Architecture of Mammalian Sperm Flagella and Implications for Ciliary Motility. EMBO J. 2021, 40, e107410. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, D.; Takeda, H. Ciliary Motility: The Components and Cytoplasmic Preassembly Mechanisms of the Axonemal Dyneins. Differentiation 2012, 83, S23–S29. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T. Axoneme Structure from Motile Cilia. Cold Spring Harb. Perspect. Biol. 2017, 9, a028076. [Google Scholar] [CrossRef] [PubMed]
- Setchell, B.P. Domestication and Reproduction. Anim. Reprod. Sci. 1992, 28, 195–202. [Google Scholar] [CrossRef]
- Skjæraasen, J.; Mayer, I.; Meager, J.; Rudolfsen, G.; Karlsen, Ø.; Haugland, T.; Kleven, O. Sperm Characteristics and Competitive Ability in Farmed and Wild Cod. Mar. Ecol. Prog. Ser. 2009, 375, 219–228. [Google Scholar] [CrossRef]
- Maes, D.; Lopez Rodriguez, A.; Rijsselaere, T.; Vyt, P.; Van Soom, A. Artificial Insemination in Pigs. In Artificial Insemination in Farm Animals; Manafi, M., Ed.; InTech: Houston, TX, USA, 2011; ISBN 978-953-307-312-5. [Google Scholar]
- Lopez Rodriguez, A.; Van Soom, A.; Arsenakis, I.; Maes, D. Boar Management and Semen Handling Factors Affect the Quality of Boar Extended Semen. Porc. Health Manag. 2017, 3, 15. [Google Scholar] [CrossRef]
- Gerrits, R.; Lunney, J.; Johnson, A.; Pursel, V.; Kraeling, R.; Rohrer, J.; Dobrinsky, J. Perspectives for Artificial Insemination and Genomics to Improve Global Swine Populations. Theriogenology 2005, 63, 283–299. [Google Scholar] [CrossRef]
- Vyt, P. Examination and Storage of Liquid Porcine Semen. Ph.D Thesis, Ghent University, Merelbeke, Belgium, 2007. [Google Scholar]
- Maes, D.; Nauwynck, H.; Rijsselaere, T.; Mateusen, B.; Vyt, P.; De Kruif, A.; Van Soom, A. Diseases in Swine Transmitted by Artificial Insemination: An Overview. Theriogenology 2008, 70, 1337–1345. [Google Scholar] [CrossRef]
- Van Der Meer, J.R.; De Vos, W.M.; Harayama, S.; Zehnder, A.J. Molecular Mechanisms of Genetic Adaptation to Xenobiotic Compounds. Microbiol. Rev. 1992, 56, 677–694. [Google Scholar] [CrossRef]
- Leaver, M.J.; Diab, A.; Boukouvala, E.; Williams, T.D.; Chipman, J.K.; Moffat, C.F.; Robinson, C.D.; George, S.G. Hepatic Gene Expression in Flounder Chronically Exposed to Multiply Polluted Estuarine Sediment: Absence of Classical Exposure ‘Biomarker’ Signals and Induction of Inflammatory, Innate Immune and Apoptotic Pathways. Aquat. Toxicol. 2010, 96, 234–245. [Google Scholar] [CrossRef]
- Larson, G.; Albarella, U.; Dobney, K.; Rowley-Conwy, P.; Schibler, J.; Tresset, A.; Vigne, J.-D.; Edwards, C.J.; Schlumbaum, A.; Dinu, A.; et al. Ancient DNA, Pig Domestication, and the Spread of the Neolithic into Europe. Proc. Natl. Acad. Sci. USA 2007, 104, 15276–15281. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.R.; Neff, N.F.; Kalisky, T.; Dalerba, P.; Treutlein, B.; Rothenberg, M.E.; Mburu, F.M.; Mantalas, G.L.; Sim, S.; Clarke, M.F.; et al. Quantitative Assessment of Single-Cell RNA-Sequencing Methods. Nat. Methods 2014, 11, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Griffith, O.L.; Mwenifumbo, J.; Goya, R.; Morrissy, A.S.; Morin, R.D.; Corbett, R.; Tang, M.J.; Hou, Y.-C.; Pugh, T.J.; et al. Alternative Expression Analysis by RNA Sequencing. Nat. Methods 2010, 7, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.R. “Validation” in Genome-Scale Research. J. Biol. 2008, 8, 3. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-Sequencing Analysis Workflows Using Whole-Transcriptome RT-qPCR Expression Data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef]
- Coenye, T. Do Results Obtained with RNA-Sequencing Require Independent Verification? Biofilm 2021, 3, 100043. [Google Scholar] [CrossRef]
- Fulgione, D.; Maselli, V.; Rivieccio, E.; Aceto, S.; Salvemini, M.; Buglione, M. Evolutionary Plasticity in Insular Lizard, Adapting over Reproduction, Metabolism, and Color Variation. Biology 2023, 12, 1478. [Google Scholar] [CrossRef]
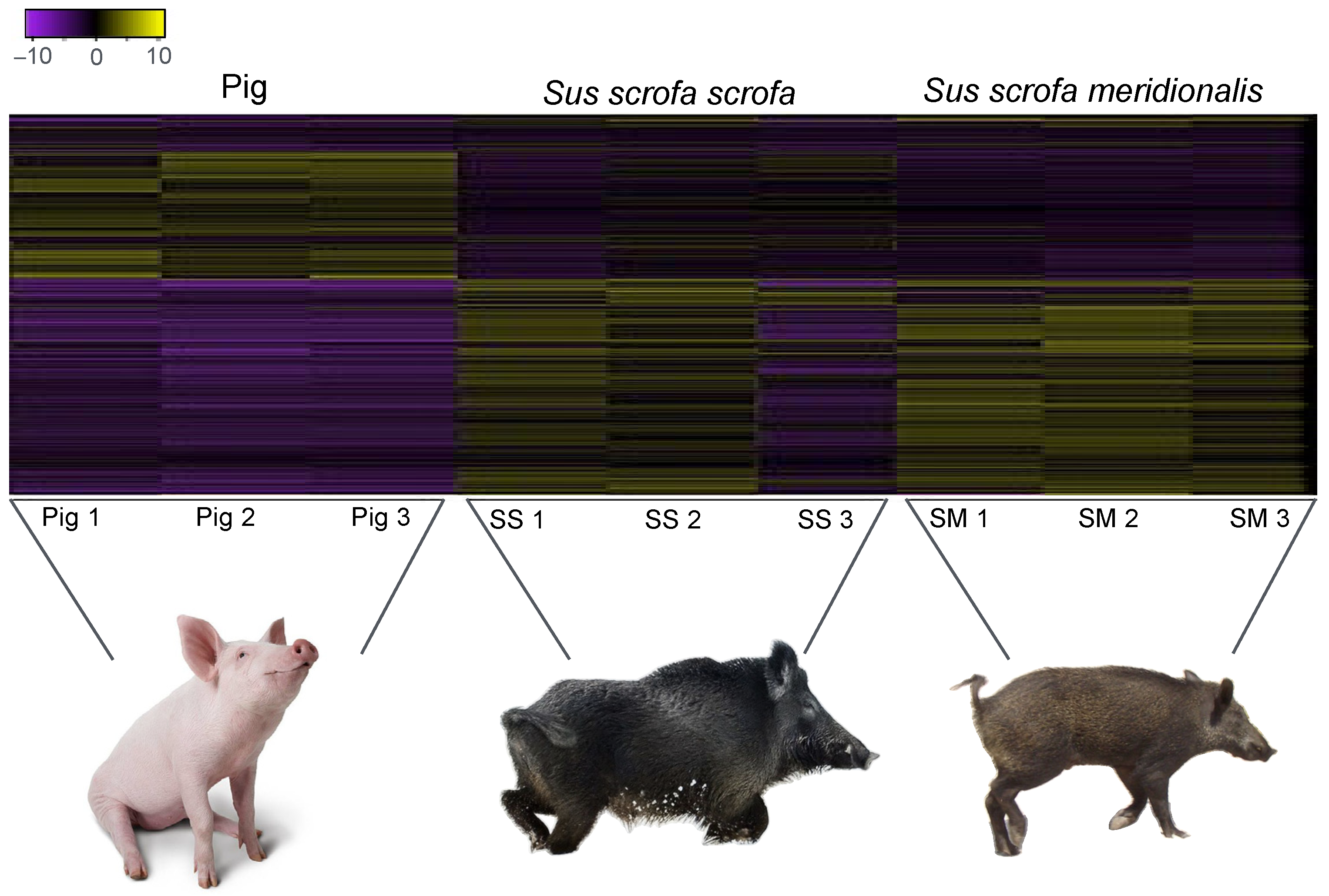
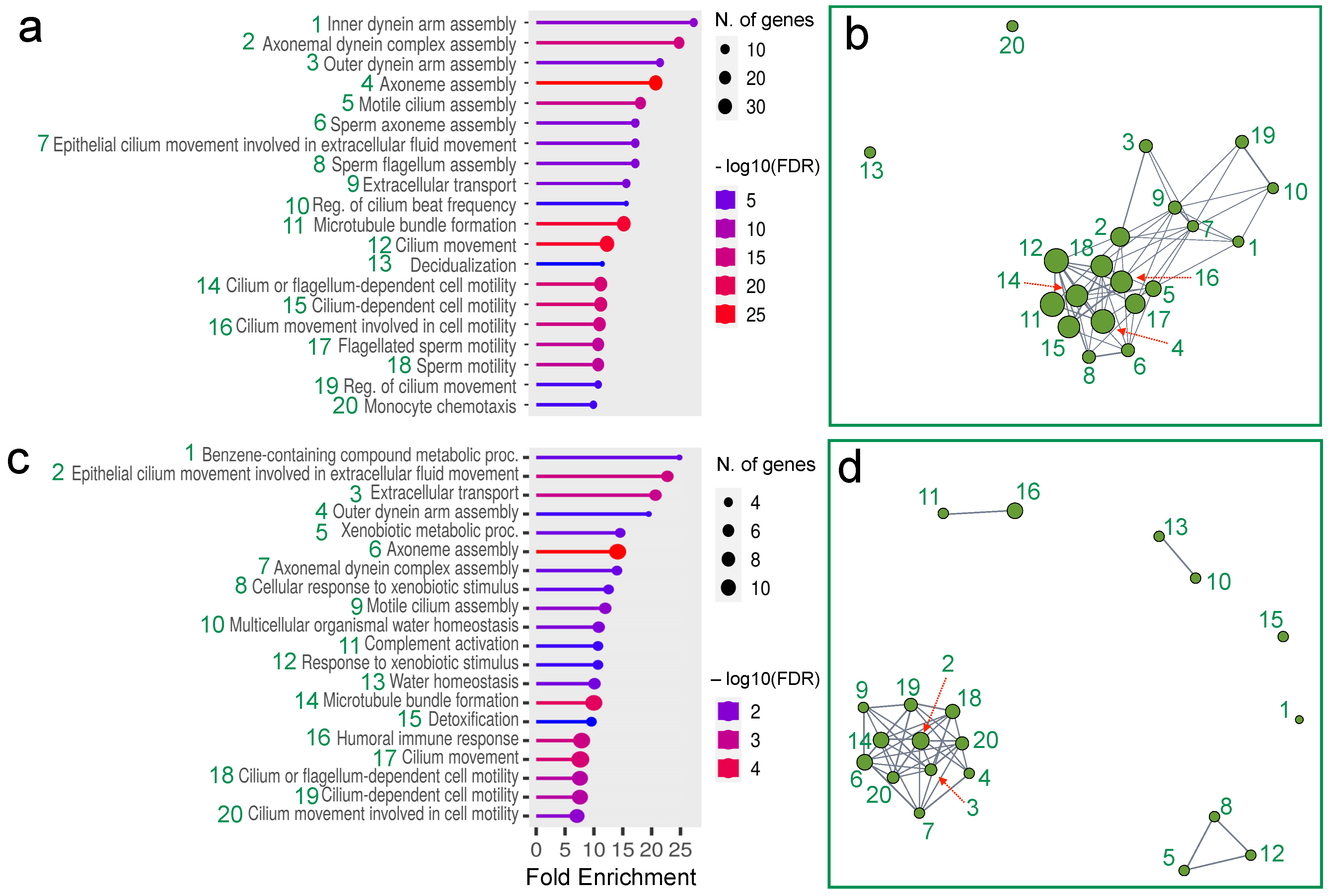

| Sample | #Input Reads | %Mapped Reads | #Mapped Reads | #Mean Mapped Reads Per Type | s.d. |
|---|---|---|---|---|---|
| PIG_1 | 20,663,211 | 86.85 | 17,947,762 | 17,114,785 | 2,111,330 |
| PIG_2 | 20,909,420 | 89.35 | 18,682,567 | ||
| PIG_3 | 17,359,635 | 84.76 | 14,714,027 | ||
| SS_1 | 26,605,201 | 86.36 | 22,976,252 | 19,000,282 | 3,580,300 |
| SS_2 | 20,012,531 | 89.91 | 17,993,267 | ||
| SS_3 | 17,692,668 | 90.61 | 16,031,327 | ||
| SM_1 | 22,432,413 | 89.27 | 20,025,415 | 17,474,470 | 2,271,444 |
| SM_2 | 17,523,002 | 89.43 | 15,670,821 | ||
| SM_3 | 18,126,543 | 92.28 | 16,727,174 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buglione, M.; Rivieccio, E.; Aceto, S.; Paturzo, V.; Biondi, C.; Fulgione, D. The Domestication of Wild Boar Could Result in a Relaxed Selection for Maintaining Olfactory Capacity. Life 2024, 14, 1045. https://doi.org/10.3390/life14081045
Buglione M, Rivieccio E, Aceto S, Paturzo V, Biondi C, Fulgione D. The Domestication of Wild Boar Could Result in a Relaxed Selection for Maintaining Olfactory Capacity. Life. 2024; 14(8):1045. https://doi.org/10.3390/life14081045
Chicago/Turabian StyleBuglione, Maria, Eleonora Rivieccio, Serena Aceto, Vincenzo Paturzo, Carla Biondi, and Domenico Fulgione. 2024. "The Domestication of Wild Boar Could Result in a Relaxed Selection for Maintaining Olfactory Capacity" Life 14, no. 8: 1045. https://doi.org/10.3390/life14081045
APA StyleBuglione, M., Rivieccio, E., Aceto, S., Paturzo, V., Biondi, C., & Fulgione, D. (2024). The Domestication of Wild Boar Could Result in a Relaxed Selection for Maintaining Olfactory Capacity. Life, 14(8), 1045. https://doi.org/10.3390/life14081045





