Comparison of the Skin Microbiota in the Periocular Region between Patients with Inflammatory Skin Diseases and Healthy Participants: A Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. DNA Extraction, PCR Amplification, and 16S rRNA Gene Sequencing
2.3. Bioinformatics and Statistical Analysis
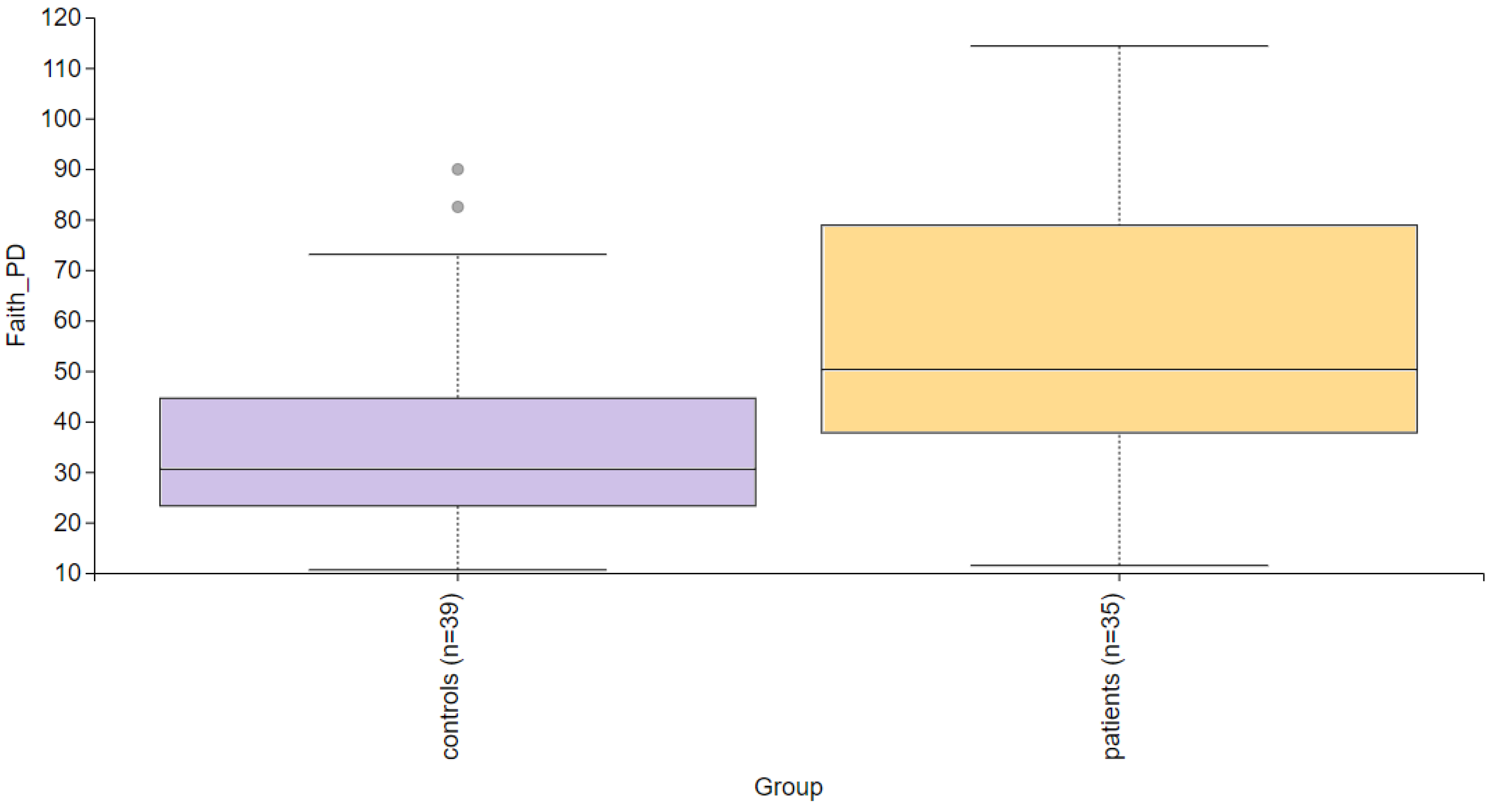
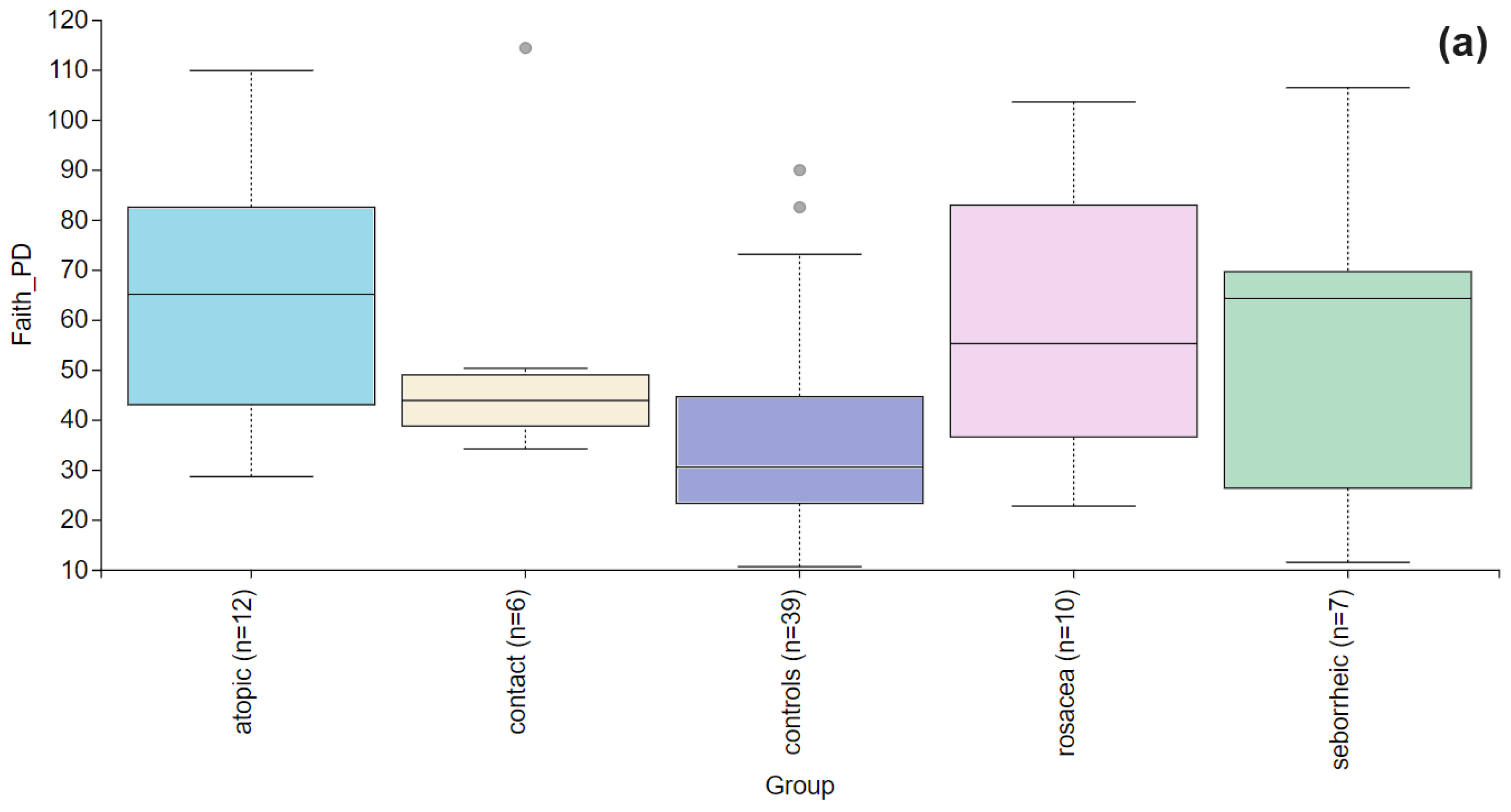
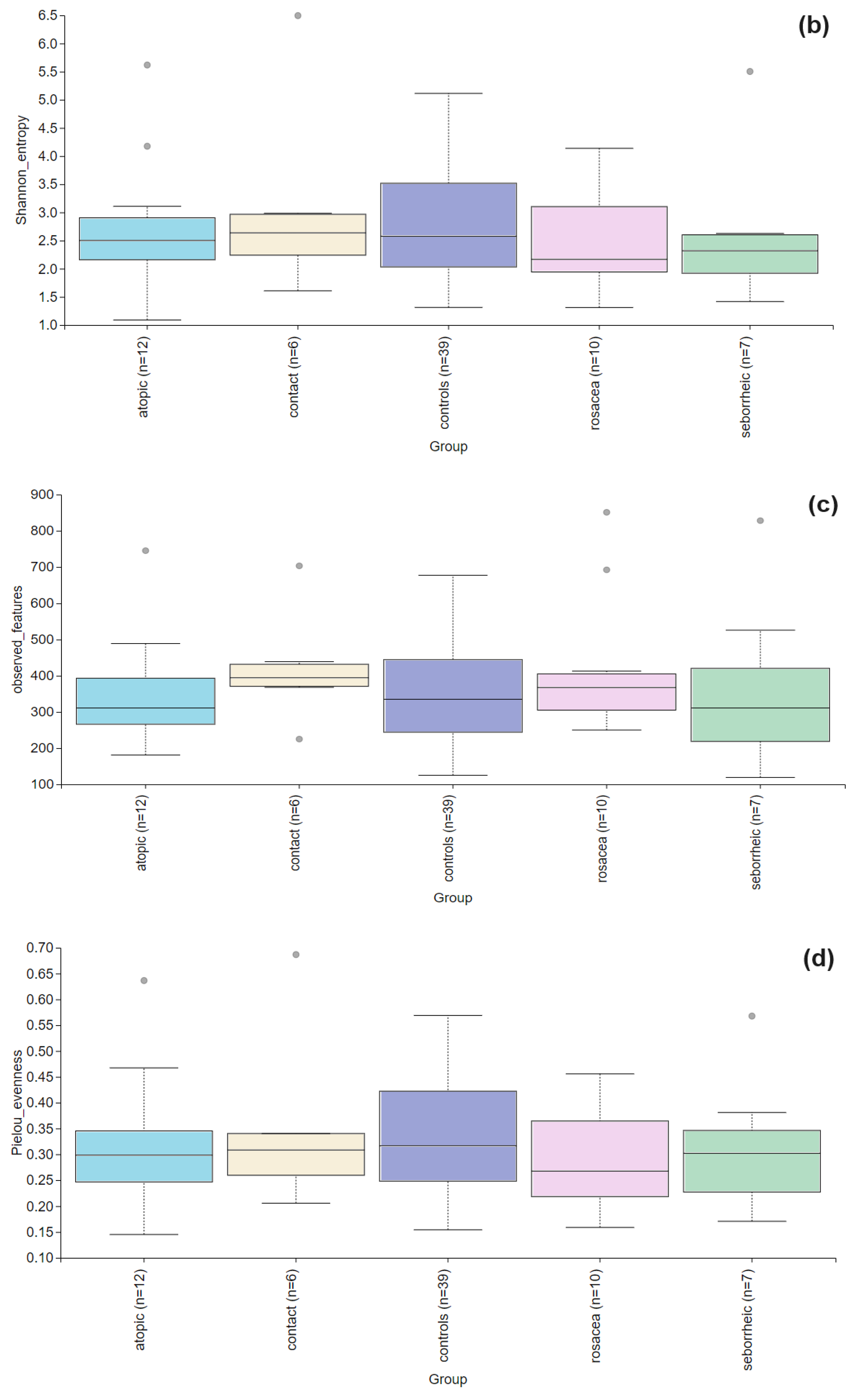
3. Results
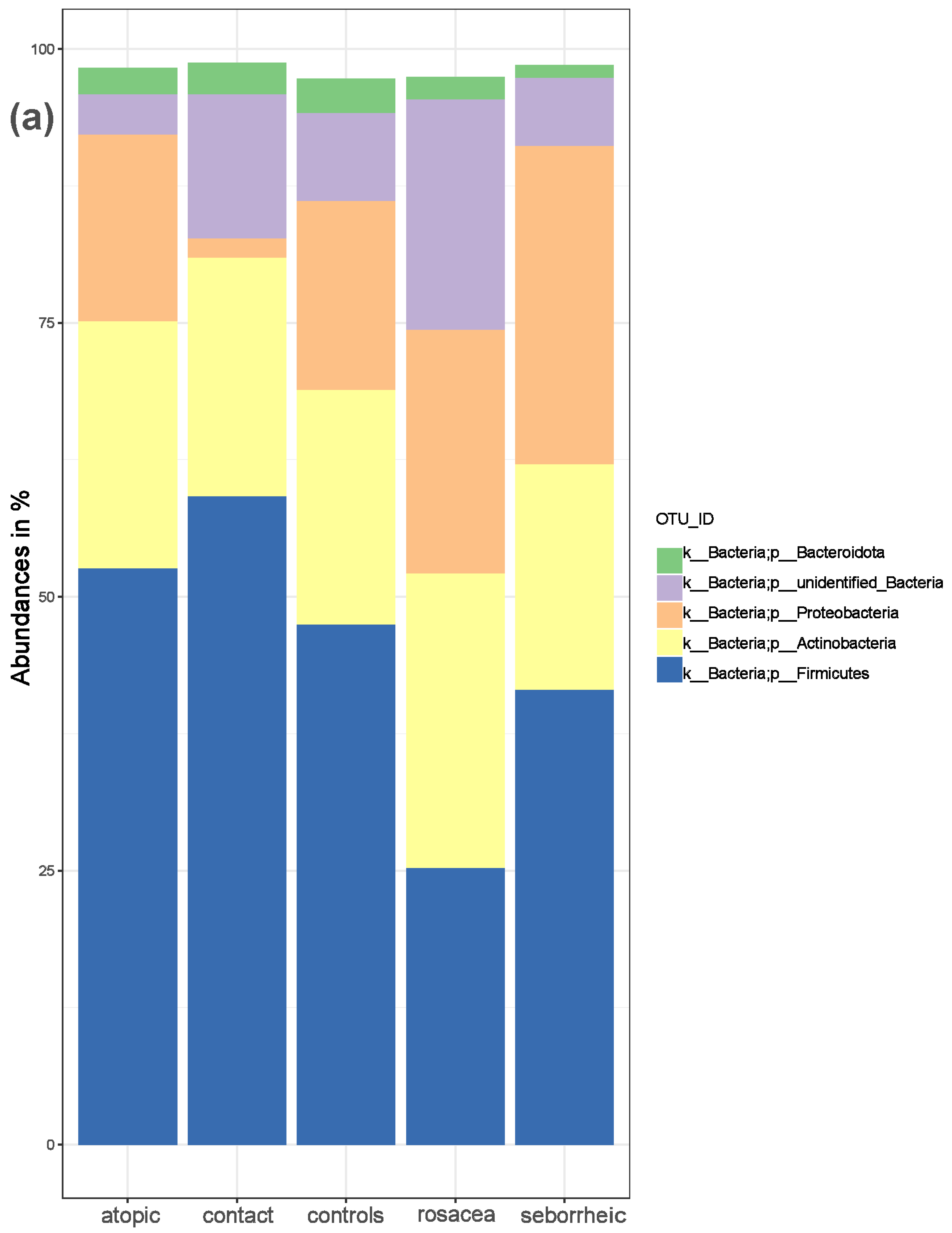
3.1. Composition of Bacterial Skin Microbiota in PD Patients and Healthy Controls
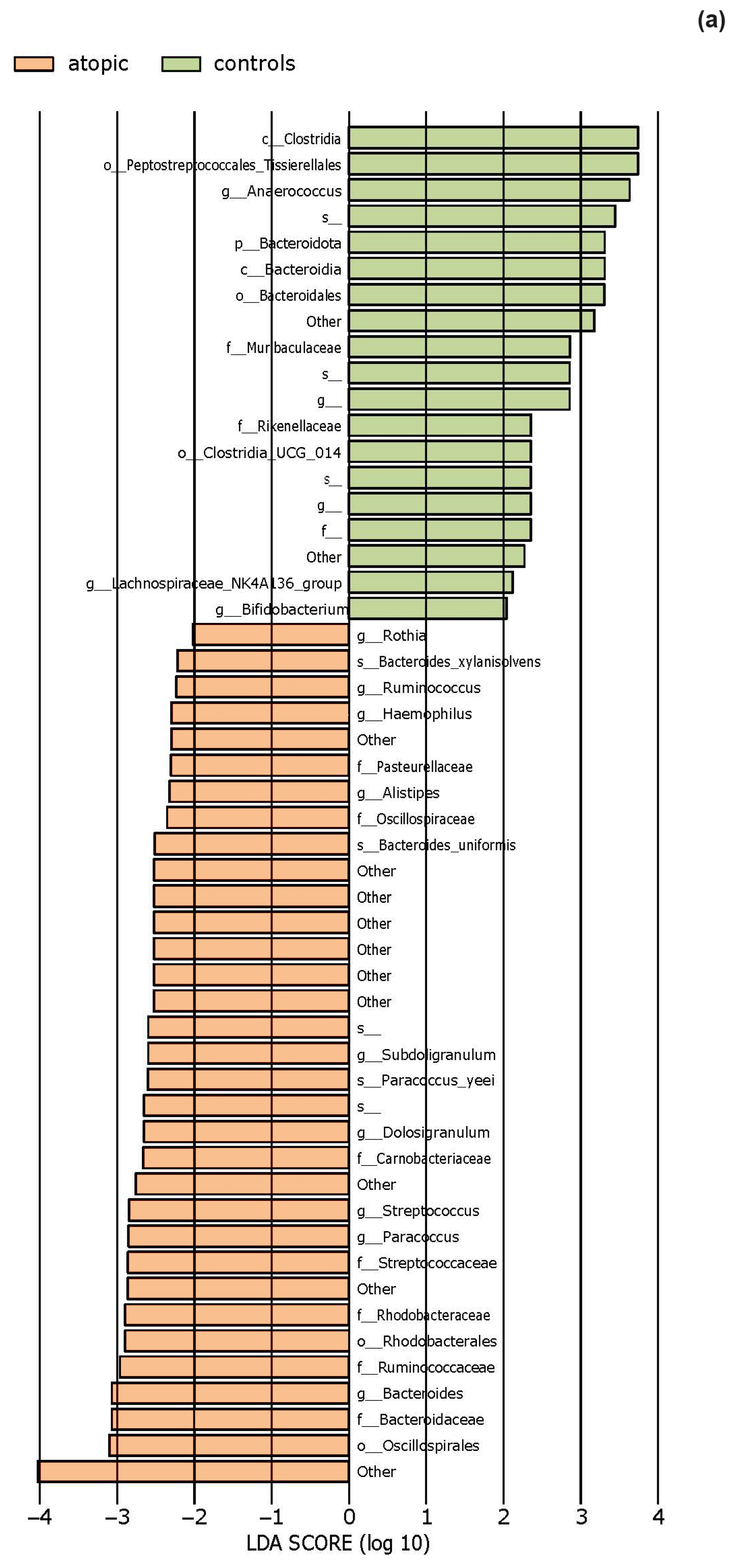
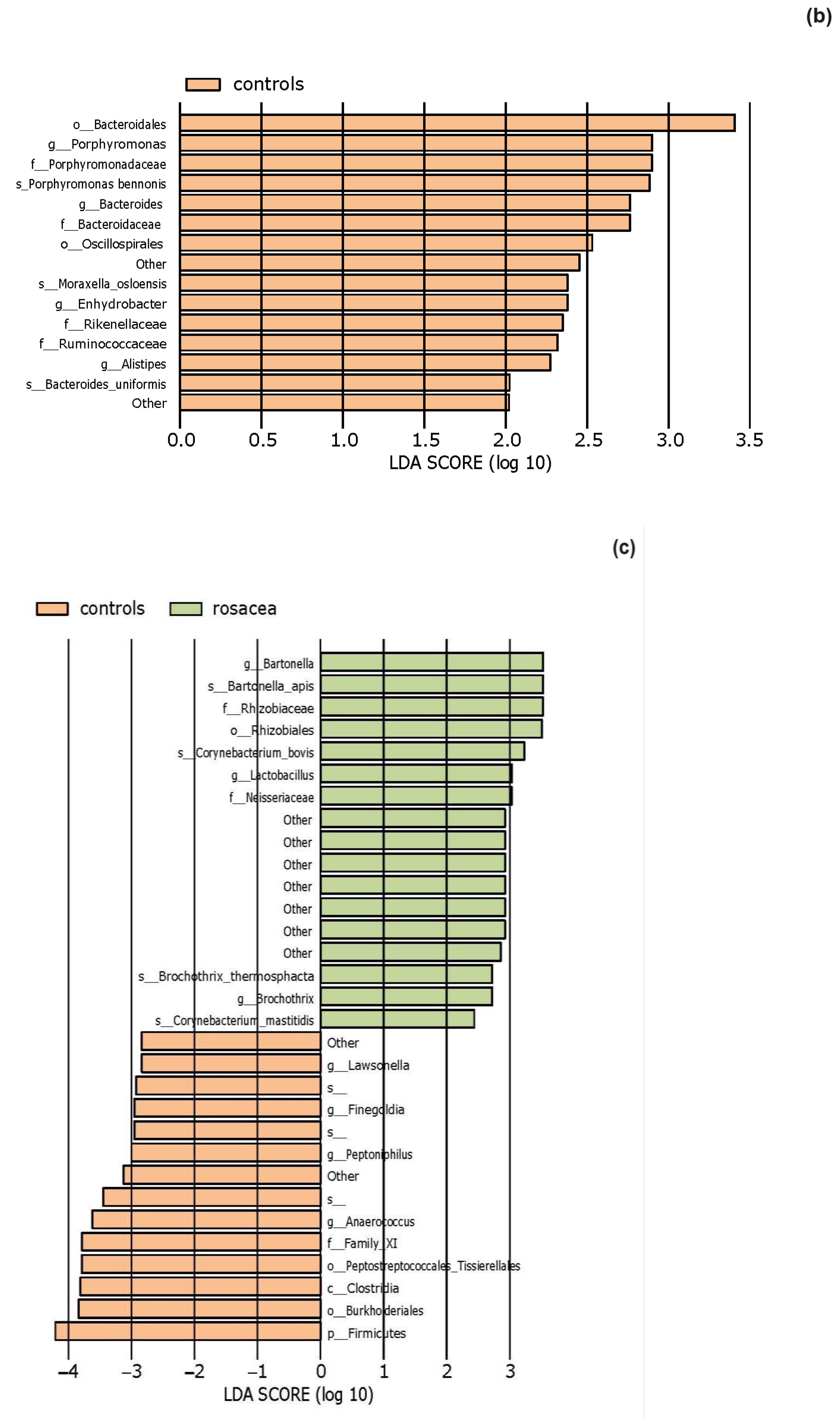
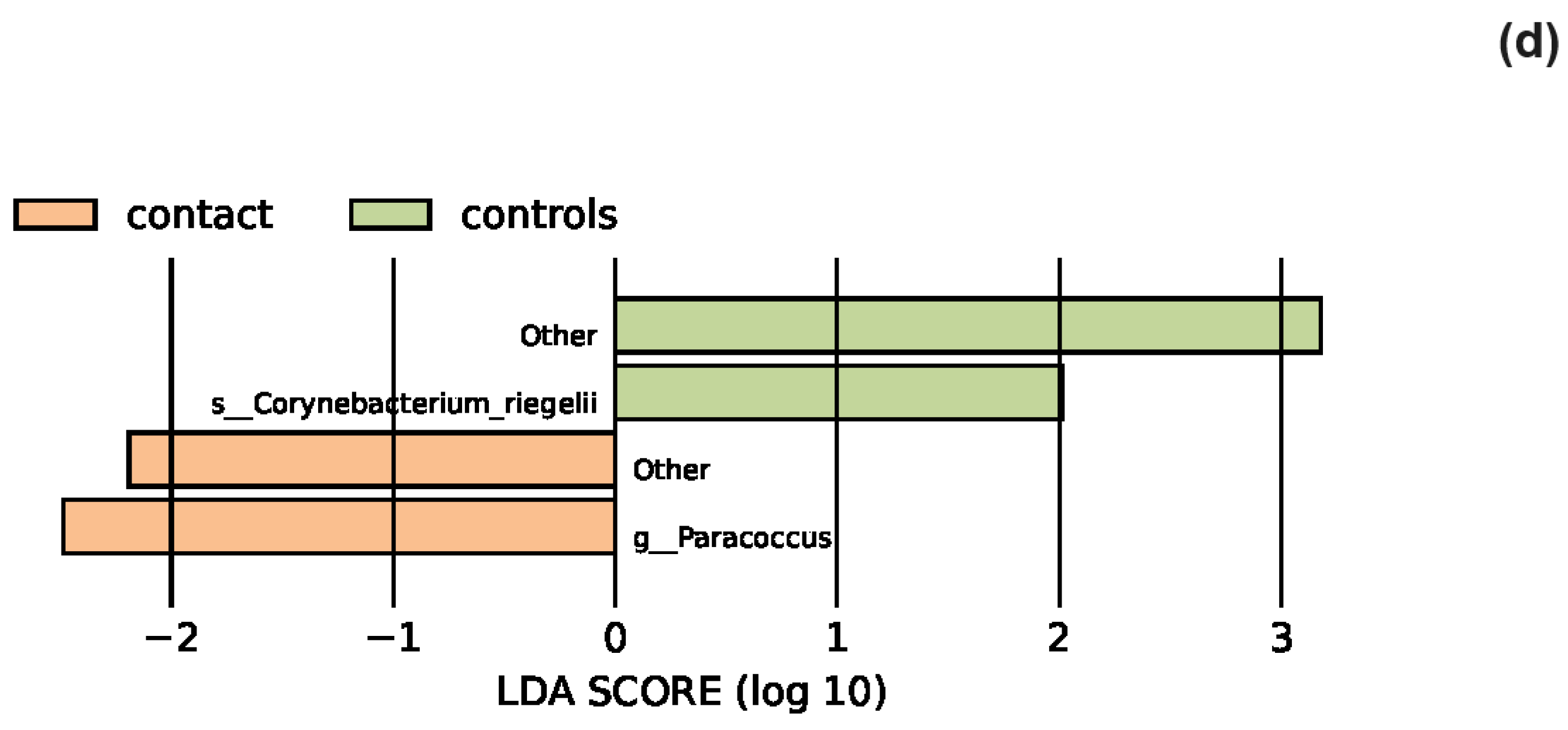
3.2. The Statistical Analysis of the Most Abundant Taxonomic Groups
4. Discussion
4.1. Characteristics of the Skin Microbiota in Patients with Atopic Dermatitis
4.2. Characteristics of the Skin Microbiota in Patients with Seborrheic Dermatitis
4.3. Characteristics of the Skin Microbiota in Patients with Rosacea
4.4. Characteristics of the Skin Microbiota in Patients with Contact Dermatitis
4.5. Study Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolf, R.; Orion, E.; Tüzün, Y. Periorbital (eyelid) dermatides. Clin. Dermatol. 2014, 32, 131–140. [Google Scholar] [CrossRef]
- Chang, P.; Moreno-Coutiño, G. Periocular dermatoses. Int. J. Womens Dermatol. 2017, 3, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Feser, A.; Plaza, T.; Vogelgsang, L.; Mahler, V. Periorbital dermatitis—A recalcitrant disease: Causes and differential diagnoses. Br. J. Dermatol. 2008, 159, 858–863. [Google Scholar] [CrossRef]
- Landeck, L.; Schalock, P.C.; Baden, L.A.; Gonzalez, E. Periorbital contact sensitization. Am. J. Ophthalmol. 2010, 150, 366–370.e2. [Google Scholar] [CrossRef]
- Crouse, L.; Ziemer, C.; Ziemer, C.; Lugo-Somolinos, A. Trends in Eyelid Dermatitis. Dermatitis 2018, 29, 96–97. [Google Scholar] [CrossRef] [PubMed]
- Guin, J.D. Eyelid dermatitis: Experience in 203 cases. J. Am. Acad. Dermatol. 2002, 47, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Ayala, F.; Fabbrocini, G.; Bacchilega, R.; Berardesca, E.; Caraffini, S.; Corazza, M.; Flori, M.L.; Francalanci, S.; Guarrera, M.; Lisi, P.; et al. Gruppo Italiano di Ricerca sulle Dermatiti da Contatto e Ambientali della Società Italiana di Dermatologia e Venereologia. Eyelid dermatitis: An evaluation of 447 patients. Am. J. Contact Dermat. 2003, 14, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Warshaw, E.M.; Voller, L.M.; Maibach, H.I.; Zug, K.A.; DeKoven, J.G.; Atwater, A.R.; Reeder, M.J.; Sasseville, D.; Taylor, J.S.; Fowler, J.F., Jr.; et al. Eyelid dermatitis in patients referred for patch testing: Retrospective analysis of North American Contact Dermatitis Group data, 1994–2016. J. Am. Acad. Dermatol. 2021, 84, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Cavuoto, K.M.; Mendez, R.; Miller, D.; Galor, A.; Banerjee, S. Effect of clinical parameters on the ocular surface miccrobiome in children and adults. Clin. Ophthalmol. 2018, 12, 1189–1197. [Google Scholar] [CrossRef]
- Kim, M.; Jang, H.; Rho, S. Risk factors for periorbital dermatitis in patients using dorzolamide/timolol eye drops. Sci. Rep. 2021, 11, 17896. [Google Scholar] [CrossRef]
- Slominski, A.; Wortsman, J.; Luger, T.; Paus, R.; Solomon, S. Corticotropin releasing hormone and proopiomelanocortin involvement in the cutaneous response to stress. Physiol. Rev. 2000, 80, 979–1020. [Google Scholar] [CrossRef]
- Ferček, I.; Lugović-Mihić, L.; Tambić-Andrašević, A.; Ćesić, D.; Grginić, A.G.; Bešlić, I.; Mravak-Stipetić, M.; Mihatov-Štefanović, I.; Buntić, A.M.; Čivljak, R. Features of the Skin Microbiota in Common Inflammatory Skin Diseases. Life 2021, 11, 962. [Google Scholar] [CrossRef]
- Paller, A.S.; Kong, H.H.; Seed, P.; Naik, S.; Scharschmidt, T.C.; Gallo, R.L.; Luger, T.; Irvine, A.D. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.E.; Zheng, P.; Ye, S.Z.; Ma, X.; Liu, E.; Pang, Y.B.; He, Q.Y.; Zhang, Y.X.; Li, W.Q.; Zeng, J.H.; et al. Microbiome: Role in Inflammatory Skin Diseases. J. Inflamm. Res. 2024, 17, 1057–1082. [Google Scholar] [CrossRef] [PubMed]
- Langan, E.A.; Griffiths, C.E.M.; Solbach, W.; Knobloch, J.K.; Zillikens, D.; Thaçi, D. The role of the microbiome in psoriasis: Moving from disease description to treatment selection? Br. J. Dermatol. 2018, 178, 1020–1027. [Google Scholar] [CrossRef]
- Scher, J.U. The Microbiome in Psoriasis and Psoriatic Arthritis: Joints. J. Rheumatol. Suppl. 2018, 94, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Li, R.; Wang, R. Skin microbiome alterations in seborrheic dermatitis and dandruff: A systematic review. Exp. Dermatol. 2021, 30, 1546–1553. [Google Scholar] [CrossRef]
- Tutka, K.; Żychowska, M.; Żaczek, A.; Maternia-Dudzik, K.; Pawełczyk, J.; Strapagiel, D.; Lach, J.; Reich, A. Skin Microbiome in Prurigo Nodularis. Int. J. Mol. Sci. 2023, 24, 7675. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Smythe, P.; Wilkinson, H.N. The Skin Microbiome: Current Landscape and Future Opportunities. Int. J. Mol. Sci. 2023, 24, 3950. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Ujiie, H.; Rosmarin, D.; Schön, M.P.; Ständer, S.; Boch, K.; Metz, M.; Maurer, M.; Thaci, D.; Schmidt, E.; Cole, C.; et al. Unmet Medical Needs in Chronic, Non-communicable Inflammatory Skin Diseases. Front. Med. 2022, 9, 875492. [Google Scholar] [CrossRef] [PubMed]
- Pesqué, D.; Aerts, O.; Bizjak, M.; Gonçalo, M.; Dugonik, A.; Simon, D.; Ljubojević-Hadzavdić, S.; Malinauskiene, L.; Wilkinson, M.; Czarnecka-Operacz, M.; et al. Differential diagnosis of contact dermatitis: A practical-approach review by the EADV Task Force on contact dermatitis. J. Eur. Acad. Dermatol. Venereol. 2024, epub ahead of print. [CrossRef] [PubMed]
- Suzuki, T.; Sutani, T.; Nakai, H.; Shirahige, K.; Kinoshita, S. The Microbiome of the Meibum and Ocular Surface in Healthy Subjects. Investig. Ophthalmol. Vis. Sci. 2020, 61, 18. [Google Scholar] [CrossRef]
- Brandwein, M.; Fuks, G.; Israel, A.; Sabbah, F.; Hodak, E.; Szitenberg, A.; Harari, M.; Steinberg, D.; Bentwich, Z.; Shental, N.; et al. Skin Microbiome Compositional Changes in Atopic Dermatitis Accompany Dead Sea Climatotherapy. Photochem. Photobiol. 2020, 96, 450. [Google Scholar] [CrossRef]
- Edslev, S.M.; Olesen, C.M.; Nørreslet, L.B.; Ingham, A.C.; Iversen, S.; Lilje, B.; Clausen, M.L.; Jensen, J.S.; Stegger, M.; Agner, T.; et al. Staphylococcal Communities on Skin Are Associated with Atopic Dermatitis and Disease Severity. Microorganisms 2021, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Bjerre, R.D.; Holm, J.B.; Palleja, A.; Sølberg, J.; Skov, L.; Johansen, J.D. Skin dysbiosis in the microbiome in atopic dermatitis is site-specific and involves bacteria, fungus and virus. BMC Microbiol. 2021, 21, 256. [Google Scholar] [CrossRef]
- Tao, R.; Wang, R.; Wan, Z.; Song, Y.; Wu, Y.; Li, R. Ketoconazole 2% cream alters the skin fungal microbiome in seborrheic dermatitis: A cohort study. Clin. Exp. Dermatol. 2022, 47, 1088–1096. [Google Scholar] [CrossRef]
- Rainer, B.M.; Thompson, K.G.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Bui, J.; Fischer, A.H.; Pasieka, H.B.; Garza, L.A.; Kang, S.; et al. Characterization and Analysis of the Skin Microbiota in Rosacea: A Case-Control Study. Am. J. Clin. Dermatol. 2020, 21, 139–147. [Google Scholar] [CrossRef]
- Thompson, K.G.; Rainer, B.M.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Kang, S.; Chien, A.L. Comparison of the skin microbiota in acne and rosacea. Exp. Dermatol. 2021, 30, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Farhat, M.; Na, J.; Li, R.; Wu, Y. Bacterial and fungal microbiome characterization in patients with rosacea and healthy controls. Br. J. Dermatol. 2020, 183, 1112–1114. [Google Scholar] [CrossRef]
- Zaidi, A.K.; Spaunhurst, K.; Sprockett, D.; Thomason, Y.; Mann, M.W.; Fu, P.; Ammons, C.; Gerstenblith, M.; Tuttle, M.S.; Popkin, D.L. Characterization of the facial microbiome in twins discordant for rosacea. Exp. Dermatol. 2018, 27, 295–298. [Google Scholar] [CrossRef]
- Kim, M.H.; Rho, M.; Choi, J.P.; Choi, H.I.; Park, H.K.; Song, W.J.; Min, T.K.; Cho, S.H.; Cho, Y.J.; Kim, Y.K.; et al. A Metagenomic Analysis Provides a Culture-Independent Pathogen Detection for Atopic Dermatitis. Allergy Asthma Immunol. Res. 2017, 9, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Suwarsa, O.; Hazari, M.N.; Dharmadji, H.P.; Dwiyana, R.F.; Effendi, R.M.R.A.; Hidayah, R.M.N.; Avriyanti, E.; Gunawan, H.; Sutedja, E. A Pilot Study: Composition and Diversity of 16S rRNA Based Skin Bacterial Microbiome in Indonesian Atopic Dermatitis Population. Clin. Cosmet. Investig. Dermatol. 2021, 14, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar] [CrossRef] [PubMed]
- Zeeuwen, P.L.; Ederveen, T.H.; van der Krieken, D.A.; Niehues, H.; Boekhorst, J.; Kezic, S.; Hanssen, D.A.; Otero, M.E.; van Vlijmen-Willems, I.M.; Rodijk-Olthuis, D.; et al. Gram-positive anaerobe cocci are underrepresented in the microbiome of filaggrin-deficient human skin. J. Allergy Clin. Immunol. 2017, 139, 1368–1371. [Google Scholar] [CrossRef]
- Murdoch, D.A. Gram-positive anaerobic cocci. Clin. Microbiol. Rev. 1998, 11, 81–120. [Google Scholar] [CrossRef]
- van der Krieken, D.A.; Rikken, G.; Ederveen, T.H.A.; Jansen, P.A.M.; Rodijk-Olthuis, D.; Meesters, L.D.; van Vlijmen-Willems, I.M.J.J.; van Cranenbroek, B.; van der Molen, R.G.; Schalkwijk, J.; et al. Gram-positive anaerobic cocci guard skin homeostasis by regulating host-defense mechanisms. iScience 2023, 26, 106483. [Google Scholar] [CrossRef]
- Kim, H.B.; Um, J.Y.; Chung, B.Y.; Kim, J.C.; Kang, S.Y.; Park, C.W.; Kim, H.O. Aryl Hydrocarbon Receptors: Evidence of Therapeutic Targets in Chronic Inflammatory Skin Diseases. Biomedicines 2022, 10, 1087. [Google Scholar] [CrossRef]
- Kezic, S.; Kammeyer, A.; Calkoen, F.; Fluhr, J.W.; Bos, J.D. Natural moisturizing factor components in the stratum corneum as biomarkers of filaggrin genotype: Evaluation of minimally invasive methods. Br. J. Dermatol. 2009, 161, 1098–1104. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; NISC Comparative Sequence Program; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Moosbrugger-Martinz, V.; Hackl, H.; Gruber, R.; Pilecky, M.; Knabl, L.; Orth-Höller, D.; Dubrac, S. Initial Evidence of Distinguishable Bacterial and Fungal Dysbiosis in the Skin of Patients with Atopic Dermatitis or Netherton Syndrome. J. Investig. Dermatol. 2021, 141, 114–123. [Google Scholar] [CrossRef]
- Miernikiewicz, P.; Barylski, J.; Wilczak, A.; Dragoš, A.; Rybicka, I.; Bałdysz, S.; Szymczak, A.; Dogsa, I.; Rokush, K.; Harhala, M.A.; et al. New Phage-Derived Antibacterial Enzyme PolaR Targeting Rothia spp. Cells 2023, 12, 1997. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.G.H.; Nijsten, T.; Verlouw, J.; Kraaij, R.; Pardo, L.M. Composition of cutaneous bacterial microbiome in seborrheic dermatitis patients: A cross-sectional study. PLoS ONE 2021, 16, e0251136. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Panchamukhi, A.; Li, P.; Shan, W.; Zhou, H.; Hou, L.; Chen, W. Malassezia and Staphylococcus dominate scalp microbiome for seborrheic dermatitis. Bioprocess. Biosyst. Eng. 2021, 44, 965–975. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, Z.; Yuan, C.; Liu, X.; Yang, F.; Wang, T.; Wang, J.; Manabe, K.; Qin, O.; Wang, X.; et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 2016, 6, 24877. [Google Scholar] [CrossRef] [PubMed]
- Massiot, P.; Clavaud, C.; Thomas, M.; Ott, A.; Guéniche, A.; Panhard, S.; Muller, B.; Michelin, C.; Kerob, D.; Bouloc, A.; et al. Continuous clinical improvement of mild-to-moderate seborrheic dermatitis and rebalancing of the scalp microbiome using a selenium disulfide-based shampoo after an initial treatment with ketoconazole. J. Cosmet. Dermatol. 2022, 21, 2215–2225. [Google Scholar] [CrossRef]
- Clavaud, C.; Jourdain, R.; Bar-Hen, A.; Tichit, M.; Bouchier, C.; Pouradier, F.; El Rawadi, C.; Guillot, J.; Ménard-Szczebara, F.; Breton, L.; et al. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS ONE 2013, 8, e58203. [Google Scholar] [CrossRef]
- Grimshaw, S.G.; Smith, A.M.; Arnold, D.S.; Xu, E.; Hoptroff, M.; Murphy, B. The diversity and abundance of fungi and bacteria on the healthy and dandruff affected human scalp. PLoS ONE 2019, 14, e0225796. [Google Scholar] [CrossRef]
- Saxena, R.; Mittal, P.; Clavaud, C.; Dhakan, D.B.; Hegde, P.; Veeranagaiah, M.M.; Saha, S.; Souverain, L.; Roy, N.; Breton, L.; et al. Comparison of Healthy and Dandruff Scalp Microbiome Reveals the Role of Commensals in Scalp Health. Front. Cell Infect. Microbiol. 2018, 8, 346. [Google Scholar] [CrossRef]
- Rousel, J.; Saghari, M.; Pagan, L.; Nădăban, A.; Gambrah, T.; Theelen, B.; de Kam, M.L.; Haakman, J.; van der Wall, H.E.C.; Feiss, G.L.; et al. Treatment with the Topical Antimicrobial Peptide Omiganan in Mild-to-Moderate Facial Seborrheic Dermatitis versus Ketoconazole and Placebo: Results of a Randomized Controlled Proof-of-Concept Trial. Int. J. Mol. Sci. 2023, 24, 14315. [Google Scholar] [CrossRef]
- Rauf, A.; Khalil, A.A.; Rahman, U.U.; Khalid, A.; Naz, S.; Shariati, M.A.; Rebezov, M.; Urtecho, E.Z.; de Albuquerque, R.D.D.G.; Anwar, S.; et al. Recent advances in the therapeutic application of short-chain fatty acids (SCFAs): An updated review. Crit. Rev. Food Sci. Nutr. 2022, 62, 6034–6054. [Google Scholar] [CrossRef]
- Polak, K.; Bergler-Czop, B.; Szczepanek, M.; Wojciechowska, K.; Frątczak, A.; Kiss, N. Psoriasis and Gut Microbiome-Current State of Art. Int. J. Mol. Sci. 2021, 22, 4529. [Google Scholar] [CrossRef] [PubMed]
- Ćesić, D.; Lugović Mihić, L.; Ozretić, P.; Lojkić, I.; Buljan, M.; Šitum, M.; Zovak, M.; Vidović, D.; Mijić, A.; Galić, N.; et al. Association of Gut Lachnospiraceae and Chronic Spontaneous Urticaria. Life 2023, 13, 1280. [Google Scholar] [CrossRef]
- Xiao, X.; Hu, X.; Yao, J.; Cao, W.; Zou, Z.; Wang, L.; Qin, H.; Zhong, D.; Li, Y.; Xue, P.; et al. The role of short-chain fatty acids in inflammatory skin diseases. Front. Microbiol. 2023, 13, 1083432. [Google Scholar] [CrossRef]
- Abdel-Gadir, A.; Stephen-Victor, E.; Gerber, G.K.; Noval Rivas, M.; Wang, S.; Harb, H.; Wang, L.; Li, N.; Crestani, E.; Spielman, S.; et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat. Med. 2019, 25, 1164–1174. [Google Scholar] [CrossRef]
- Liu, R.; Peng, C.; Jing, D.; Xiao, Y.; Zhu, W.; Zhao, S.; Zhang, J.; Chen, X.; Li, J. Biomarkers of Gut Microbiota in Chronic Spontaneous Urticaria and Symptomatic Dermographism. Front. Cell Infect. Microbiol. 2021, 11, 703126. [Google Scholar] [CrossRef]
- Almoughrabie, S.; Cau, L.; Cavagnero, K.; O’Neill, A.M.; Li, F.; Roso-Mares, A.; Mainzer, C.; Closs, B.; Kolar, M.J.; Williams, K.J.; et al. Commensal Cutibacterium acnes induce epidermal lipid synthesis important for skin barrier function. Sci. Adv. 2023, 9, eadg6262. [Google Scholar] [CrossRef]
- Woo, Y.R.; Lee, S.H.; Cho, S.H.; Lee, J.D.; Kim, H.S. Characterization and Analysis of the Skin Microbiota in Rosacea: Impact of Systemic Antibiotics. J. Clin. Med. 2020, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Clanner-Engelshofen, B.M.; French, L.E.; Reinholz, M. Corynebacterium kroppenstedtii subsp. demodicis is the endobacterium of Demodex folliculorum. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1043–1049. [Google Scholar] [CrossRef]
- Forton, F.M.N. The Pathogenic Role of Demodex Mites in Rosacea: A Potential Therapeutic Target Already in Erythematotelangiectatic Rosacea? Dermatol. Ther. 2020, 10, 1229–1253. [Google Scholar] [CrossRef]
- Maluki, A.; Breitschwerdt, E.; Bemis, L.; Greenberg, R.; Mozayeni, B.R.; Dencklau, J.; Ericson, M. Imaging analysis of Bartonella species in the skin using single-photon and multi-photon (second harmonic generation) laser scanning microscopy. Clin. Case Rep. 2020, 8, 1564–1570. [Google Scholar] [CrossRef]
- Murillo, N.; Mediannikov, O.; Aubert, J.; Raoult, D. Bartonella quintana detection in Demodex from erythematotelangiectatic rosacea patients. Int. J. Infect. Dis. 2014, 29, 176–177. [Google Scholar] [CrossRef][Green Version]
- Boodman, C.; Gupta, N.; van Griensven, J.; Van Bortel, W. Bartonella quintana detection among arthropods and their hosts: A systematic review and meta-analysis. Parasit. Vectors 2024, 17, 328. [Google Scholar] [CrossRef] [PubMed]
- Rainer, B.M.; Mongodin, E.; Bui, J.; Fischer, A.; Pasieka, H.; Garza, L.A.; Kang, S.; Chien, A. Finegoldia magna and Corynebacterium kroppenstedtii are significantly enriched in rosacea independent of rosacea subtype: Results of a case-control study. J. Investig. Dermatol. 2017, 137, S265. [Google Scholar] [CrossRef][Green Version]
- Masuda-Kuroki, K.; Alimohammadi, S.; Di Nardo, A.S. epidermidis Rescues Allergic Contact Dermatitis in Sphingosine 1-Phosphate Receptor 2-Deficient Skin. Int. J. Mol. Sci. 2023, 24, 13190. [Google Scholar] [CrossRef] [PubMed]
- Igawa, S.; Ohzono, A.; Pham, P.; Wang, Z.; Nakatsuji, T.; Dokoshi, T.; Di Nardo, A. Sphingosine 1-Phosphate Receptor 2 Is Central to Maintaining Epidermal Barrier Homeostasis. J. Investig. Dermatol. 2021, 141, 1188–1197.e5. [Google Scholar] [CrossRef]


| Variable | Patients | Controls | p-Value | |
|---|---|---|---|---|
| All patients | N | 35 | 39 | |
| Age, median (range) [years] | 58 (22–86) | 60 (23–88) | 0.455 | |
| Sex, n (%) | ||||
| Women | 26 (74.29) | 23 (59.0) | 0.220 | |
| Men | 9 (25.71) | 16 (41.0) | ||
| Rosacea | n (%) | 10 (28.57) | ||
| Age, median (range) [years] | 57.5 (36–74) | 0.503 | ||
| Sex, n (%) | ||||
| Women | 8 (80.00) | 0.288 | ||
| Men | 2 (20.00) | |||
| Seborrheic dermatitis | n (%) | 7 (20.00) | ||
| Age, median (range) [years] | 73 (61–82) | 0.095 | ||
| Sex, n (%) | ||||
| Women | 4 (57.14) | 1.000 | ||
| Men | 3 (42.86) | |||
| Atopic dermatitis | n (%) | 12 (34.29) | ||
| Age, median (range) [years] | 37.5 (22–86) | 0.023 | ||
| Sex, n (%) | ||||
| Women | 10 (83.33) | 0.174 | ||
| Men | 2 (16.67) | |||
| Contact dermatitis | n (%) | 6 (17.14) | ||
| Age, median (range) [years] | 61.5 (38–82) | 0.841 | ||
| Sex, n (%) | ||||
| Women | 4 (66.67) | 1.000 | ||
| Men | 2 (33.33) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferček, I.; Ozretić, P.; Tambić-Andrašević, A.; Trajanoski, S.; Ćesić, D.; Jelić, M.; Geber, G.; Žaja, O.; Paić, J.; Lugović-Mihić, L.; et al. Comparison of the Skin Microbiota in the Periocular Region between Patients with Inflammatory Skin Diseases and Healthy Participants: A Preliminary Study. Life 2024, 14, 1091. https://doi.org/10.3390/life14091091
Ferček I, Ozretić P, Tambić-Andrašević A, Trajanoski S, Ćesić D, Jelić M, Geber G, Žaja O, Paić J, Lugović-Mihić L, et al. Comparison of the Skin Microbiota in the Periocular Region between Patients with Inflammatory Skin Diseases and Healthy Participants: A Preliminary Study. Life. 2024; 14(9):1091. https://doi.org/10.3390/life14091091
Chicago/Turabian StyleFerček, Iva, Petar Ozretić, Arjana Tambić-Andrašević, Slave Trajanoski, Diana Ćesić, Marko Jelić, Goran Geber, Orjena Žaja, Josipa Paić, Liborija Lugović-Mihić, and et al. 2024. "Comparison of the Skin Microbiota in the Periocular Region between Patients with Inflammatory Skin Diseases and Healthy Participants: A Preliminary Study" Life 14, no. 9: 1091. https://doi.org/10.3390/life14091091
APA StyleFerček, I., Ozretić, P., Tambić-Andrašević, A., Trajanoski, S., Ćesić, D., Jelić, M., Geber, G., Žaja, O., Paić, J., Lugović-Mihić, L., & Čivljak, R. (2024). Comparison of the Skin Microbiota in the Periocular Region between Patients with Inflammatory Skin Diseases and Healthy Participants: A Preliminary Study. Life, 14(9), 1091. https://doi.org/10.3390/life14091091











