The molecular translation machine consists of various parts and accessories, such as ribozymes, amino acids, tRNAs, aaRS, mRNAs, ribozymes, peptides, and various enzymes. In the modern translation machine, mRNA is decoded in a ribosome to produce a specific amino acid chain or polypeptide. The polypeptide then folds into an active protein and performs its function in the cell. The list of parts of translation machine is not sufficient condition for understanding its biologic function, such as programmed protein synthesis. Understanding how the parts work in unison is also important. However, it is not enough. We have to do reverse engineering to reconstruct how these parts might have evolved and interacted in the prebiotic environment. The origin and evolution of the translation machine may shed new light on how the information system emerged in the peptide/RNA world.
In the peptide/RNA world, peptides played significant roles in accelerating the chemical reactions, by lowering activation energy. Some long peptides are good catalysts and show some enzymic activity. Most likely, ten proteinogenic amino acids were abiotically synthesized [
67]. These ancestral amino acids gave rise to a limited variety of random peptides and polypeptides; most were useless, without much specificity, but a few were specifically selected for their catalytic activity. The need for both specific and a wide range of protein enzymes became essential in the peptide/RNA world for biogenesis. Peptides are distinguished from proteins on the basis of size and origin. The peptide is short (only few amino acids long), the protein is long (more than ~40 amino acids), folded, and it forms the catalytic center with a fixed start and end. As a result, the protein enzyme is much more versatile for catalytic reaction than the primitive peptide. Peptides and polypeptides could abiotically form in the prebiotic environment, but the proteins could not arise by chance but are coded, because it is achieved after a long evolutionary process. The evolution of peptides to proteins occurred from a small motif of short peptides to longer folded peptides, and finally to proteins that form complex catalytic centers with almost unlimited possible functions [
24]. Darwinian selection provided the driving force for the evolution of specific protein over peptide, so that protein synthesis became essential to the protocell function. These coded proteins were custom-made by translation machines consisting of a repertoire of RNA and protein molecules, and they were highly specified for protocellular functions. The evolution of protein was a long evolutionary process that was driven by incremental advances of the translation machinery, which facilitated the transition from random, simple, peptide produced through an abiotic process, to the eventual production of specific, complex, proteins by RNA-directed protein synthesis.
The evolution of complex information system must consist of plausible, elementary steps, with each conferring a distinct advantage on the evolving ensemble of genetic elements. Here, we map the emergence of potential informational and catalytic oligomers, derived from the assembly of building blocks, and reconstruct the probable steps that lead to the translation machinery and the genetic code. It is well-known that modern protein synthesis proceeds with the participation of 20 amino acids, ribozymes, tRNA, various enzymes, including aminoacyl tRNA synthetase (aaRS), mRNA, ribosomal RNA, ribosomal proteins, ribosome, a considerable number of proteinous factors, ATP, GTP, etc. More than 120 species of RNAs and proteins are involved in the process of protein synthesis [
65]. The most important steps include: base pair complementarity, the origin of ribozyme, the origin of tRNA, the origin of aminoacyl-tRNA synthetase, the origin of mRNA, the origin of ribosome, the synthesis of protein, and the origin of the genetic code and translation.
We identify nine major stages for the origin and evolution of the translation machinery complex and the genetic code leading to the protein synthesis. These possible biochemical pathways are: (1) the selection of amino acids; (2) the origin of RNA; (3) the origin of ribozyme; (4) the origin of transfer RNA; (5) the origin of metabolism; (6) the origin of aminoacyl-tRNA synthetase; (7) the origin of messenger RNA and translation; (8) the origin of ribosome; and finally, (9) protein synthesis. During the emergence of these biochemical pathways, the genetic code and the translation system coevolved with the translation machine.
5.2. The Origin of RNA
The basic constituents of RNA molecules, such as D-ribose, phosphate, and the four bases—adenine (A), guanine (G), cytosine (C), and uracil (U), along with unused nucleotides, were delivered to the hydrothermal crater lake by meteorites [
2,
3,
4]. The polymerization of RNA molecules occurred by mineral catalysis in the prebiotic environment. Nucleotide monomers were linked on the montmorillonite clay substrates of the crater floor in an ATP-rich environment [
1,
9,
22]. The accumulation of phosphates in the vent environment was an important requirement in making the sugar-phosphate backbone of RNA. Nucleotides underwent spontaneous polymerization on the mineral substrate with the loss of water. The resulting product was a mixture of polynucleotides that were random in length and sequence.
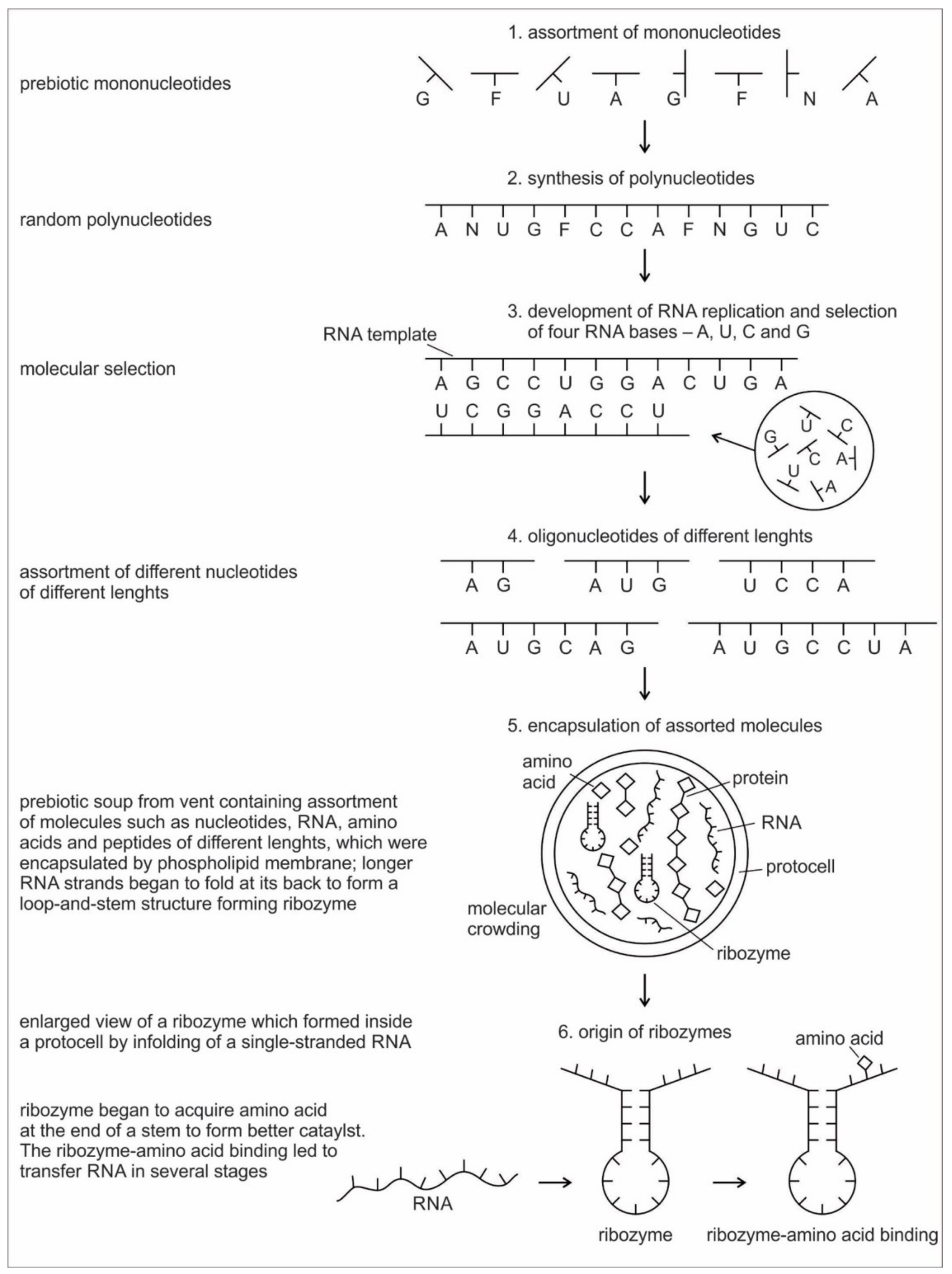
Six hypothetical stages for the formation of the RNA molecules in the prebiotic environment is shown in
Figure 2. It seems unlikely that the prebiotic soup in the prebiotic environment produced only the four bases that were found in RNA—A, U, G, and C—which formed the polynucleotide chain. Certainly, there were other nucleotides (including hypothetical F and N bases in
Figure 2), which were incapable of Watson–Crick base pairing. Initially, all of these mononucleotides were randomly polymerized into short oligonucleotides of different lengths by peptide bonding [
40]. The process was mediated by natural selection and RNA replication. Natural selection led to the elimination of useless random oligonucleotide sequences during base pairing. From these chaotic assemblages of oligonucleotides, only four bases, such as A, U, C, and G, were selected by exploiting the properties of the Watson–Crick base pairing, whereas hypothetical F and N bases were eliminated. The four standard bases are better than 2 or 6 based on estimates of arbitrary catalysis and the actual pairing energy of standard bases [
45]. The four nucleotides were strung together to produce short pieces of oligonucleotide and RNA molecules, which could replicate with the aid of the peptide enzyme. Replication selected prebiotic RNA molecular bases from overwhelmingly large assortment of mononucleotides. These RNA molecules were random and noncoded, being a jumble assortment of nucleotide bases.
Once some rudimentary template-dependent synthetic mechanism allowing for base-pairing was in place, molecules rich in A, U, C, and G were then progressively selected and amplified. These bases joined to form primordial RNA strands of different lengths, which began to self-replicate through a process of base pairing. Short sequences of nucleotides are normally better replicators than long sequences. Longer sequences suffer from an important evolutionary disadvantage; it takes longer to replicate a long sequence than a short once. If a pool of nucleotide sequences containing a range of length is left to code and replicate, then short sequences will dominate and long ones will become extinct [
70]. The base pairing principle would later give rise to codon-anticodon hybridization, the origin of messenger RNA, transcription, and replication.
RNA is generally single-stranded and an informational molecule. The self-replication of RNA molecules occurs through a process of base pairing and dissociation. When one RNA strand is made in the vent environment, a second strand would automatically form through base pairing in such a way that cytosine always pairs with guanine, while adenine always pairs with uracil. Consequently, pairing is always between purine and pyrimidine. Because the hydrothermal vents in the crater basins were hot, double-stranded RNA, which formed by base-pairing, came apart through the dissociation of the two chains. When the strands separate, the cycle repeats with another round of base pairing, leading to two more double-stranded RNA molecules, one of which contains the original strand, containing its exact copy. By exploiting the properties of nucleotide base-pairing, coupled with the high temperatures of hydrothermal vent in the crater basin, short pieces of RNA replicated without the aid of any other molecules. Such complementary templating mechanisms lie at the heart of RNA replication, producing a large, more diverse population of RNA molecules (
Figure 2).
Because RNA contains a sequence of bases that is analogous to the letters in a word, it can function as an information containing molecule. Moreover, RNA, being a single chain, is free to take any kind of shape; the structure that it can achieve by morphing its shape is wide-ranging, similar to protein. From this basic architecture of a single-stranded RNA molecule, different species of RNAs, such as ribozymes, tRNA, mRNA, and rRNA evolved inside protocells, with a supply of information, distinct in attribute and configuration in response to amino acids. There was a molecular choreography of different RNAs in the prebiotic world that led to the rudimentary translation. The advent and multifunction of different species of RNA molecules signal the transition from the age of chemistry to the age of information.
5.3. The Origin of Ribozyme
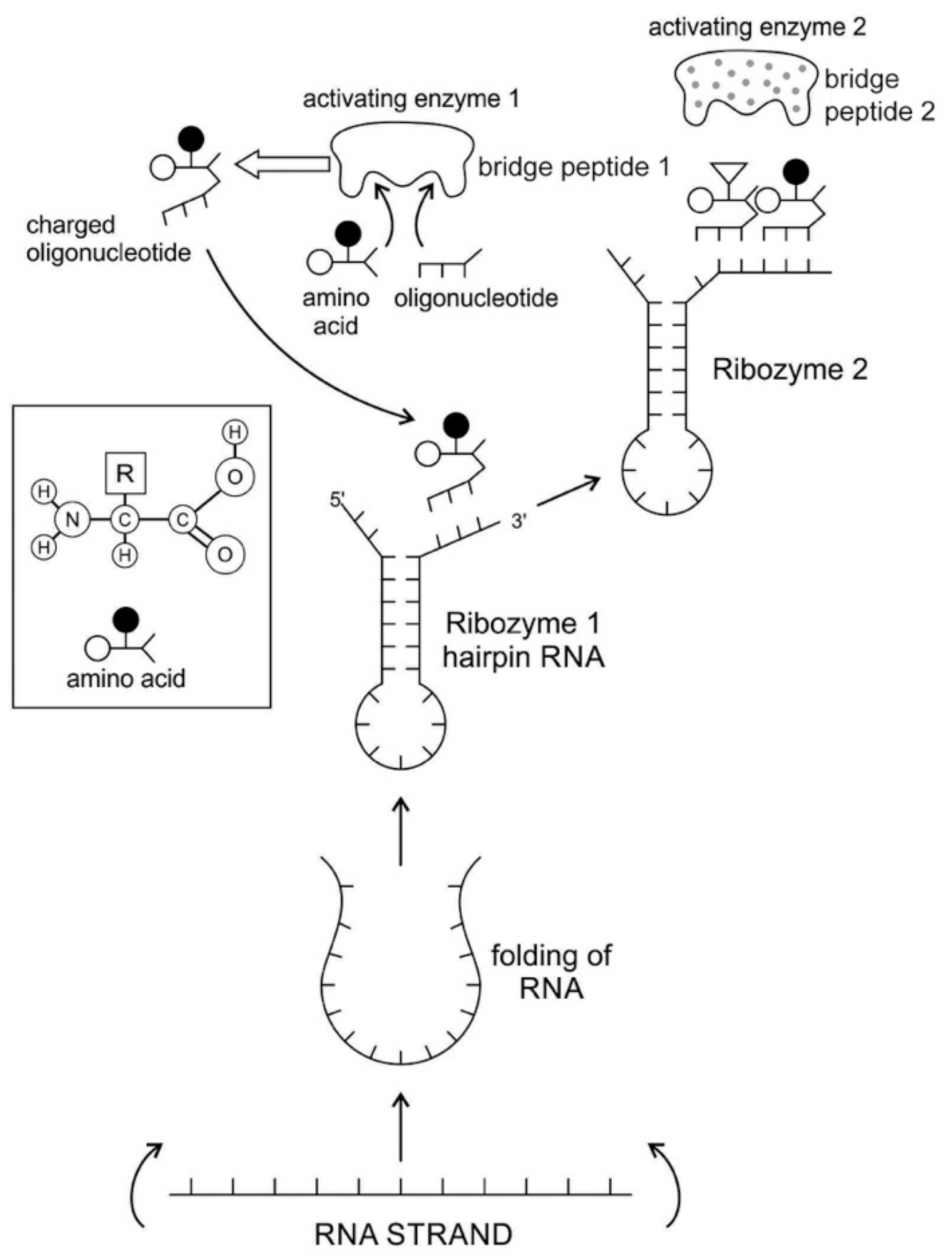
The RNA molecule has a secondary structure. It can form a localized double-stranded RNA stem by base pairing and a terminal loop to form a hairpin structure. In the stem, adenine forms a bond with uracil and cytosine pairs with guanine to form double-stranded RNA. The resulting hairpin structure is a key building block of many RNA secondary structures, such as ribozyme and tRNA (
Figure 3). As an important secondary structure of RNA, an RNA hairpin can direct RNA folding, determine interactions in a ribozyme, protect structural stability for mRNA, provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reaction [
71]. Structurally, RNA hairpins can occur in different positions within different types of RNAs; they differ in the length of the stem, the size of the loop, the number and size of the bulges, and in the actual nucleotide sequence.
Ribozymes are RNA molecules that are capable of catalyzing specific biochemical reaction, similar to the action of protein enzymes. There are different classes of ribozymes, but they all appear to be associated with metal ions, such as potassium or magnesium. Different ribozymes catalyze different reactions, but almost all ribozymes are involved in catalyzing the cleavage of RNA chains in the formation of bonds between the RNA strands.
Most likely, the chemical bonding of a particular amino acid to a small RNA hairpin structure led to the origin of ribozyme. We assume that different kinds of RNA, protein enzymes, nucleotides, oligonucleotides, and amino acids were available in the prebiotic soup. The single-stranded nature of RNA molecule can be bent back on itself, in a hairpin loop, where the stems of the loops are maintained by base pairing to form a three-dimensional structure, just like a protein molecule to act as an enzyme. In some stem-loop configurations, two ends of the stem might remain free, containing the 3’ and 5’ ends. This 3’ end might function as an acceptor stem to form a covalent attachment to a specific amino acid (
Figure 3). This small hairpin RNA molecule with specific terminal base sequences acquired the corresponding amino acid as a ‘cofactor’ to improve the catalytic range and efficiency to become initial ribozymes [
46]. Many enzymes act with the help of one or more cofactors. The binding of amino acids to a ribozyme resulted in an enhancement of catalytic activity.
Any specific binding between two molecules involves information, as if two molecules ‘recognize’ each other. An amino acid can be linked to an oligonucleotide with three bases by an activating enzyme; the charged oligonucleotide is then bound on the surface of a ribozyme by base pairing and delivers the appropriate amino acid (
Figure 2). In this way, ribozymes are capable of producing short peptide chain. This de-novo peptide would play a role in stabilization, in order to become coded. Overtime, the original peptide forming ribozymes will specialize as amino acid specific adaptors. In the peptide/RNA world, different kinds of peptides were synthesized. Aminoacylation of ribozymes would be governed by the availability of amino acids. Most primordial amino acids in the prebiotic environment were alanine, glycine, valine, and aspartic acid. Initially, one kind of an amino acid and one kind of hairpin would be catalyzed by an activating enzyme, perhaps a precursor to the aminoacyl transfer tRNA synthetase, such as bridge peptide, which is a very short peptide that facilitated the emergence of self-sustained RNA-peptide complex supporting a primitive translation [
24]. The BP was initially synthesized by chance as a result of the physical proximity of hybridized short, random aminoacylated ribozyme. Each BP is somewhat specific for an amino acid and for its corresponding ribozyme, but the specificity is low. Aminoacylated ribozyme would be involved in complex formation, bringing some of the aminoacylated ribozyme 3’-ends in close proximity. This would promote peptide bond formation between two adjacent amino acids (
Figure 3). There was a feedback between ribozymes and bridge peptides. Later, a second amino acid, which is attached to a different hairpin by a different ribozyme, would be added, and so on to create a chain of polypeptide, supporting a primitive proto-translation. It is our contention that the interacting union of a hairpin ribozyme with a specific amino acid is cornerstone in the origin of information, transfer RNA, translation, genetic code, and protein synthesis. The ribozyme would give rise to tRNA and bridge peptide to pre-aaRS to aaRS. This is a combination of model between the one Koonin-developed model [
72] describing a polymer transition out of the RNA world and the model of Carter [
17] for the role of aaRS in the initial stages of code formation and translation.
A ribozyme has a well-defined tertiary structure that enables it to act like a protein enzyme in catalyzing biochemical and metabolic reactions. The relevance of ribozyme for the origin of tRNA is enormous. Ribozymes, being assembled in the prebiotic vent environment, could not only replicate themselves but would catalyze the formation of specific proteins. The adaptor ribozymes are the precursors of tRNA molecules and they play critical roles in the building of ribosomes. Ribosomal RNA functions as a peptidyl transferase in ribosomes to link the amino acids in protein synthesis, but the framework of transferase is provided by the ribosomal proteins.
5.4. The Origin of Transfer RNA
Any model for the development of protein synthesis must necessarily start with direct interactions between RNAs and amino acids. Chemical considerations suggested that direct interactions between the amino acids and the codons in mRNA were unlikely. The protein and mRNA languages seem to be unrelated. Amino acids do not read their codons. Some kind of an adaptor molecule must mediate the specification of amino acids by codons in mRNAs during protein synthesis [
27]. The adaptor molecules were soon identified by other researchers as transfer RNAs (tRNAs), which serve as a reading device of mRNA through base pairing. The tRNA molecule binds to amino acids, associates with mRNA molecules, and also interacts with ribosomes to decipher and translate the code of mRNA.
It is generally believed that the first RNA gene, the
Ur-Gen, was a precursor of modern tRNA [
73]. tRNA is the ancestor of all RNAs. It is an ancient molecule that has evolved very little over time. The phylogeny of ribosomes suggests that tRNA is an ancient component of ribosomes that arose in the early prebiotic world [
20].
A tRNA molecule is short, typically being 76 to 90 nucleotides in length, which serves as the physical link, a cipher, between the messenger RNA (mRNA) and the amino acid sequences of proteins [
30]. Although the tRNA molecule is short, both its primary structure and its overall geometry are undoubtedly more complex than those of any other RNA species [
74]. The translation of a message carried in mRNA into the amino acid language of proteins requires an interpreter. The amino acids themselves cannot recognize the codons in mRNA. The tRNA matches appropriate amino acids to the appropriate codons. To convert the three-letters words (codons) of nucleic acids to the one-letter, amino acids of proteins, tRNA molecules serves as the interpreters during translation. Each amino acid is joined to the correct tRNA by a special enzyme, aminoacyl-tRNA synthetase (aaRS).
tRNA participates in two clearly distinct steps in the translation process. The first step comprises the reactions that lead to the charging of the tRNA molecule with an amino acid. The second step comprises the complex reactions in which tRNA transfers its amino acids into a growing protein chain, in response to a specific codon. The chemical reaction catalyzed by the tRNA is simple—the joining of amino acids through peptide linkages. It performs the remarkable task of choosing the appropriate amino acid to be added to the growing protein chain by reading successive mRNA codons. The actual step of translation from mRNA into protein language occurs when amino acids and tRNAs are matched and joined. The translators that do this job are the aminoacyl-tRNA synthetases (aaRS). These enzymes are the only bilingual elements in the cell: they can recognize both the amino acid and the corresponding tRNAs. They are the key element of translation, being the links between the worlds of proteins and nucleic acids. The activation of tRNA occurs when a synthetase uses energy from ATP hydrolysis to attach an amino acid to a specific tRNA. There are twenty such synthetases, one for each amino acid. Together, they make up the complete dictionary for protein synthesis in a cryptic form that relies on tRNAs for decoding into the anticodon language. Each type of amino acid can be attached to only one type of tRNA, so each type of organism has many types of tRNA and more than 20 amino acids. There might be a coevolutionary process in which the anticodons and the corresponding amino acids were progressively mediated by natural selection. As ribosomes appear, tRNAs transport amino acids to ribosomes, where the amino acids are assembled into proteins.
Because of its molecular complexity, the origin of tRNA is controversial. The modern tRNA structure, with its complex configuration and multiple functions, might have originated from a simpler form, such as pre-tRNA molecules to select specific abiotic amino acids in the vent environment (
Figure 4A–D). The pre-tRNA molecules with hairpin structures (stem and loop) might have evolved in some evolutionary stages of protein synthesis, originating from a linear chain of RNA [
75]. The tRNA has a secondary and tertiary structure. In solution, the secondary structure of tRNA resembles a cloverleaf with three hairpin loops (
Figure 4E,F). One of these hairpin loops contains a sequence of three nucleotides, called the anticodon, which forms base pairs with the mRNA codon. The other two loops of the cloverleaf form a D-arm and a T-arm. The unlooped stem contains the free 3´ and 5´ ends of the chain. The CCA sequence at the 3´ end of the acceptor stem forms a covalent attachment to the amino acid that corresponds to the anticodon sequence. The CCA sequence of the acceptor stem offered a binding site for the amino acid. The 5´ terminal contains a phosphate group. Both the anticodon and the acceptor stem sequence correlate with the role of amino acids in folded proteins [
76]. The secondary structure tRNA molecule may provide some clue as to its ancestral molecular configuration. The cloverleaf-configuration of tRNA can be derived from a folded ribozyme with a single loop and an attachment site for the amino acid at the end of a stem (
Figure 4E).
The most plausible scenario of the origin of the tRNA molecule is based on ribozymes. The chemical bonding of particular amino acids to small RNA molecules with specific base sequences was the crucial step. Perhaps the precursor of tRNA started as a simple ribozyme with a hairpin structure (
Figure 4A,B). This ribozyme acquired amino acids at its 3’ end as a ‘cofactor’ (
Figure 3): that is, an amino acid was attached to a ribozyme and made it a more efficient catalyst [
77]. By using cofactors, the range of specificity of catalytic activity could be increased. One way of attaching an amino acid to a particular point on the surface of the ribozyme is at the end of a single-stranded unlooped stem of the hairpin, which is charged and begins to bind amino acid, which enhances the catalytic function of the ribozyme. With the stabilization of the catalytic reactions, these ribozymes began to participate in the first catalytic cycles. This configuration of a ribozyme linking an amino acid at the end may be the starting point for the origin of tRNA, where the unlooped stems contain the free 3´ and 5´ ends of the chain. This amino acid attachment to ribozymes by a specific assignment enzyme first occurred to make cofactors more efficient catalysts [
46].
Aminoacylation of tRNA is an essential event in the translation system. Although, in the modern system, protein enzymes play the sole role in tRNA aminoacylation; in the primitive translation system, ribozymes could have catalyzed aminoacylation to tRNA or ancestral tRNA-like molecules. What was the catalytic function of ribozyme? If it was attaching an amino acid to its own end, it would not be logical that the substrate amino acid is the cofactor at the same time. It has been suggested that this attachment first occurred to make cofactors and it was carried by ribozymes. The RNA world hypothesis implies that the ribozyme functioned as an assignment enzyme to attach a particular amino acid to an ancestral tRNA for aminoacylation before the emergence of aaRS [
77]. In the peptide/RNA world, we suggest that the ribozyme was not an aminoacylation catalyst; another molecule, such as bridge peptide, performed this function for the ligation of amino acid with ancestral tRNA [
24]. In the early stage of aminoacylation, pre-aaRS, originally a protein enzyme, emerged as an assignment enzyme for charging ancestral tRNA [
17,
18,
19]. In that case, the ribozyme should have another activity that is so advantageous as to help the molecule to survive. In our view, the cofactor function of ribozyme was utilized to form peptide bonds between adjacent amino acids before the emergence of the ribosome. This enzymatic activity may be precursor to that of the Peptidyl Transferase Center of the ribosome that is responsible for peptide bond formation. Another phenomenon in which the intervention of a ribozyme could have been of critical importance is RNA replication [
68].
Many studies have suggested that the modern cloverleaf structure of tRNA may have arisen from a single ancestral gene by the duplication of half-sized hairpin-like RNAs by passing through some intermediate structures [
76,
77,
78,
79,
80,
81,
82,
83]. The linkage of an amino acid with a ribozyme at the end with a hairpin loop might be the starting point for the origin of tRNA, which is a quarter size of the modern tRNA molecule [
47]. The relevance of ribozymes in the origin of tRNA is enormous. The equivalent effect of gene duplication might be accomplished by a simple ligation of two identical hairpins of folded ribozymes to create double hairpins, a D-hairpin and a T-hairpin, with an anticodon at the stem bases [
82]. RNA ligation is a powerful driving force for the emergence of tRNA, joining two hairpin loops of ribozyme (
Figure 4C). During the evolutionary transitions of the pre-tRNA molecule, the double hairpin structure with the D-hairpin and the T-hairpin formed in the ancient prebiotic world, with both the anticodon and the terminal CCA sequence adjacent to the D-hairpin (
Figure 4D) [
80].
The function of tRNA molecules depends on their precise three-dimensional structure. The cloverleaf tRNA folds into a more compact L-shaped tertiary structure, but each has a distinct anticodon and an attached amino acid (
Figure 4H). One arm of the L-shaped tRNA structure has a minihelix with a single-stranded CCA end that is used for attaching a single amino acid; the other arm forms an anticodon loop, with three unpaired bases that may bind with the complementary codon of mRNA. Each tRNA molecule can carry one of the 20 different amino acids at its CCA minihelix end. Each type of amino acid has its own type of tRNA, which binds it and carries it to the growing end of a protein chain during the decoding of mRNA. The CCA end of the minihelix interacts with the large ribosomal subunit to form a peptide bond and the loop end interacts with the small ribosomal subunit for decoding mRNA triplets through codon-anticodon interactions [
76].
We suggest that this half-sized hairpin structure of the pre-tRNA molecule acquired some functional capacity for translation before the emergence of tRNA (
Figure 4C,D). The pre-tRNA molecule is the evolutionary precursor of the tRNA molecule. Direct duplication or the ligation of half-sized, hairpin-like structures—the pre-tRNA molecule— could have formed the contemporary full-length tRNA molecules, (
Figure 4E). The acceptor stem bases and the anticodon stem/loop bases in tRNA in tRNA 5´-half and 3´-half fit together with the double-hairpin folding; this suggests that the primordial double-hairpin RNA molecules could have evolved to the structure of modern tRNA by gene duplication, with subsequent mutations to form the familiar overleaf structure [
76,
80]. In other words, two pre-tRNA molecules somehow fused together to form a tRNA molecule.
The half-sized pre-tRNA molecule with two loops (D-hairpin and T-hairpin) on one side, and anticodon and acceptor stem region of CCA end on the other side, is structurally and functionally independent and is more ancient than the other-half of the tRNA molecule [
81]. This short, self-structured strand of the pre-tRNA molecule possesses a template domain, which is chargeable through interaction with specific amino acids, is probably the predecessor of tRNA (
Figure 4C). This pre-tRNA molecule binds, with high specificity, to the amino acid corresponding to its anticodon; this reaction is catalyzed by a specific pre-aminoacyl-tRNA synthetase (pre-aaRS). tRNA evolution is closely linked to aminoacylation. There is a separate tRNA for each amino acid that carries a triplet sequences of nucleotides for anticodon. Later, the anticodon of pre-tRNA will guide the codon formation of the pre-mRNA.
It should be apparent that tRNA molecules must contain a great deal of specificity, despite their small size. Not only do they (1) have the correct anticodon sequences, so as to respond to the right codons, but they must also (2) be recognized by the correct aaRS, to be activated by the correct amino acids, and (3) bind to the appropriate sites on the ribosomes to carry out their adaptor functions.
An important aspect of the specificity between amino acids and pre-tRNA is that, once this specificity is established, a mechanism for ‘memorizing’ or encoding variations in the sequence of pre-tRNA molecules becomes possible [
73]. These pre-selected biomolecules of amino acids emerged from the existing prebiotic soup of the crater vent environments. Among the many essential components of the translation process, assignment enzymes, such as pre-aaRS, evolved to bind a specific amino acid to a pre-tRNA molecule (
Figure 4).
5.5. The Origin of Metabolism
A prebiotic origin of metabolism is not fully understood. The core structure of the metabolic pathway is very similar across all organisms, which suggests the early origin of protometabolism in the prebiotic world [
37]. Catalysts may have played an important role in establishing the early metabolism that ultimately led to the biosynthesis of protein. An intriguing possibility is that modern metabolic pathways emerged through a stepwise process of recruitment of ever more-effective catalysts to catalyze steps in primordial chemical-reaction networks. Metal ions of Fe, Mn, Zn, and Cu were also available in the vent environment, which help to mediate catalysis [
10,
11,
13]. The synthesis of small organic molecules from inorganic precursors, including mineral-mediated synthesis, is probably the stimulus for the origin of metabolism. Large Hadean impacts may have made the atmosphere transiently rich in CO, which may have played a role in the origin of life and in fueling early biological metabolism. CO was an important trace gas on the prebiotic Earth, because it has high free energy and catalyzed the key reactions of prebiotic synthesis and in fueling early biological metabolisms [
16].
Crystalline surfaces of common rock-forming minerals, such as pyrite and montmorillonite, are likely to have played several important roles in protometabolism [
21]. Mineral surfaces with well-known catalytic properties might have promoted the polymerization of monomers, such as amino acids and nucleic acids. Many of life’s essential macromolecules in the prebiotic world, including enzymes, carbohydrates, and RNA, form from water-soluble monomeric units—amino acids, sugars, and nucleic acid, respectively. Minerals surfaces provide a means to concentrate and assemble these bio-monomers. The polymerization of proteins from amino acids requires the dehydration and condensation mechanism that is precisely found in the fluctuating hydrothermal crater basins. It is well-known that amino acids concentrate and polymerize on clay minerals to form small, protein-like molecules [
84]. Such reactions occur when a solution containing amino acids evaporate in the presence of clays. Subsequent studies have shown the adsorption and polymerization of amino acids on varied crystalline surfaces [
23].
The problem of the origin of the evolution of metabolism has been recently advanced by the behavior of ZnS, which is capable of harvesting sunlight energy and converting this energy into the formation of chemical bonds of dicarboxylic acid from CO
2, thus providing the core reactions of universal metabolism before the existence of enzymes [
85]. This paper has related how prebiotic metabolites available from simple sunlight promoted reactions can catalyze the synthesis of clay minerals (i.e., a zinc clay called sauconite). The work presents an excellent example of reproductive power of clay minerals and the mechanism by which prebiotic metabolites catalyze their formation. Clay minerals that act as sponges can retain water and polar organic molecules, and they might have played a key role in concentrating and catalyzing the polymerization of key organic molecules such as RNA and protein.
Small molecules—such as amino acids, short peptides, and cofactors—may have catalyzed reactions that are required to produce more complicated organic compounds. Although their catalytic abilities are known to be limited in both acceleration and specificity as compared with later molecular RNA or protein catalysts, some small molecules are remarkably effective catalysts.
The second stage, or metabolism, defined as the first set of reactions that are catalyzed by protein enzymes (and perhaps, ribozymes) prefiguring present-day metabolism, and perhaps already including certain central systems, such as the glycolytic chain and the Krebs cycle [
68]. Centrally located within this network are the sugar phosphate reactions of glycolysis and the pentose pathway. This stage of metabolism appeared in the peptide/RNA world and it was modified and refined continuously during the origin of the first cells. As more enzymes were added and started to build their own network, new pathways could have developed.
5.6. The Origin of Aminoacyl-tRNA Synthetase
Aminoacyl-tRNA synthetases (aaRSs) are a superfamily of enzymes that are responsible for creating the pool of correctly charged aminoacyl-tRNAs, which are necessary for the translation of the genetic information (mRNA) through the ribosome. aaRSs are very ancient enzymes that are present in all organisms, and are one of the pioneer molecules that are formed by the polymerization of amino acids in incremental steps. Each enzyme catalyzes the activation of a specific amino acid and recognizes a specific tRNA for binding.
The unavailability of activated amino acids was the most critical barrier of protein synthesis. Aminoacylated ribozyme was the pioneer molecule to use the amino acid as cofactor and employed bridge peptide for activation. Later, with the development of tRNA, the activation reaction is catalyzed by specific aaRS, a derived product of bridge peptide. The first step is the formation of an aminoacyl adenylate with an amino acid and an ATP. The next step is the transfer of the aminoacyl group to a particular tRNA molecule to form aminoacyl-tRNA, or a charged tRNA. The mechanism of aaRS formation is well-known [
77]. It reveals insight into how and why the tRNA molecule creates its own bilingual enzyme aaRS that can then connect it with the appropriate amino acid. It enhances the selection and sorting of the appropriate amino acids from the prebiotic soup for protein synthesis. Each aaRS is highly specific for a given amino acid. It has a highly discriminating amino acid activation site. Both amino acids and ATP were available in the hydrothermal vent, facilitating a reaction with tRNA to form aminoacyl-tRNA synthetase. Moreover, the proofreading ability by aaRS increases the fidelity of protein synthesis.
How do aaRS choose their tRNA partners? The aaRS recognize, on the one hand, individual amino acids, which they activate via conjunction with ATP; or, aaRS activate amino acids to generate its conjugate with AMP [
77]. The synthetase first binds ATP and the corresponding amino acid to form an aminoacyl-adenylate, releasing inorganic pyrophosphate (PP
1). The next step is the transfer of the aminoacyl group of aminoacyl-AMP to a particular tRNA molecule to form aminoacyl-tRNA. The mechanism can be summarized in the following reaction series:
Thus, the equivalent of two molecules of ATP are consumed in the synthesis of each aminoacyl-tRNA. One of them is consumed in the formation of the ester linkage of aminoacyl-tRNA, whereas the other is consumed in driving the reaction forward. The activation and transfer steps for a particular amino acid are catalyzed by the same aminoacyl-tRNA synthetase. Indeed, the aminoacyl-AMP intermediate does not dissociate from the synthetase. Aminoacyl-AMP is normally a transient intermediate in the synthesis of aminoacyl-tRNA. Synthetases can recognize the anticodon loops and acceptor stems of tRNA molecules. Their precise recognition of tRNAs is as important for high-fidelity protein synthesis, as is the accurate selection of amino acids.
aaRSs come in twenty flavors, with each one being specific to an amino acid and tRNA. These twenty enzymes are widely different, each being optimized to function with its own particular amino acid and the set tRNA molecules that are appropriate to that amino acid. They can be divided into two classes, termed class I and class II. The two aaRS superfamilies evenly divide translation into ten amino acids each. The initial activating enzyme was a bridge peptide that facilitated the aminoacylation of ribozyme (
Figure 3). From bridge peptide, protozymes and then urzymes, and finally pre-aaRS and aaRS probably evolved [
17,
18,
19,
24]. We speculate that the precursor of aaRS was pre-aaRS, a hypothetical primordial ancestor that gave rise to two classes of aaRS, which are both multidomain proteins. Each aaRS uses different mechanisms of aminoacylation. In our model, the original aminoacylation enzymes were pre-aaRS, a simpler version of aaRS, which must have featured a strong linkage to the anticodon of a pre-tRNA molecule. This linkage must have featured a codon-like, trinucleotide binding site for the adaptor’s anticodon, on the pre-aaRS. We propose that pre-aaRS is an enzyme, including an anticodon, plus a domain that is capable of binding and activating an amino acid and transferring to the pre-tRNA. Pre-aaRS is analogous to ‘protozymes’ and ‘urzymes’ [
18,
19], but is somewhat more advanced, because it would allow for tRNA/anticodon recognition. Protozymes retain about 40 percent of activity of the full-length of aaRS, even though they contain only about 10 percent as many amino acids. Next came ‘urzymes’, which retain about sixty percent of activity and have the same functional repertoire as the full-length enzymes. We speculate that pre-aaRS would be as long as the urzyme, but it has acquired additional anticodon binding function. The proposed evolutionary path from bridge peptide to protozyme to urzyme to pre-aaRS to aaRS documents increases the complexity of functions and would satisfy the rule of continuity [
24].
5.7. The Origin of Messenger RNA and Translation
There are two haunting questions regarding the genesis of mRNA: (1) how mRNAs first appeared in the prebiotic environment, before the emergence of DNA and (2) how they evolved in the sequence of nucleotides, with the function of specifying amino acids as the fundamental components for the origin of the genetic code. The primordial mRNA was lost long ago in the
information stage of biogenesis, leaving no trace of its origin. While existing evidence suggests that the genetic code was influenced by physico-chemical interactions between individual amino acids and strings of nucleic acids [
69,
86], researchers have yet to piece together the stepwise mechanisms by which it evolved over time.
In the prebiotic world, different species of RNA evolved through cooperation, each with a different function. Although random RNA strands grew during prebiotic synthesis by base pairing, in which some portions of the strand might show codon-like arrangement of nucleobases, they did not contain any genetic information (
Figure 2). Moreover, the strings of nucleotide may be haphazardly interrupted by stop and start signals. A fundamental property of protein synthesis is that the amino acids are not added in a haphazard fashion. Their sequence is rigorously imposed by mRNA, which is itself is incrementally formed by tRNA. Each mRNA must be specially made allowing hybridization with tRNA, and specific to each protein.
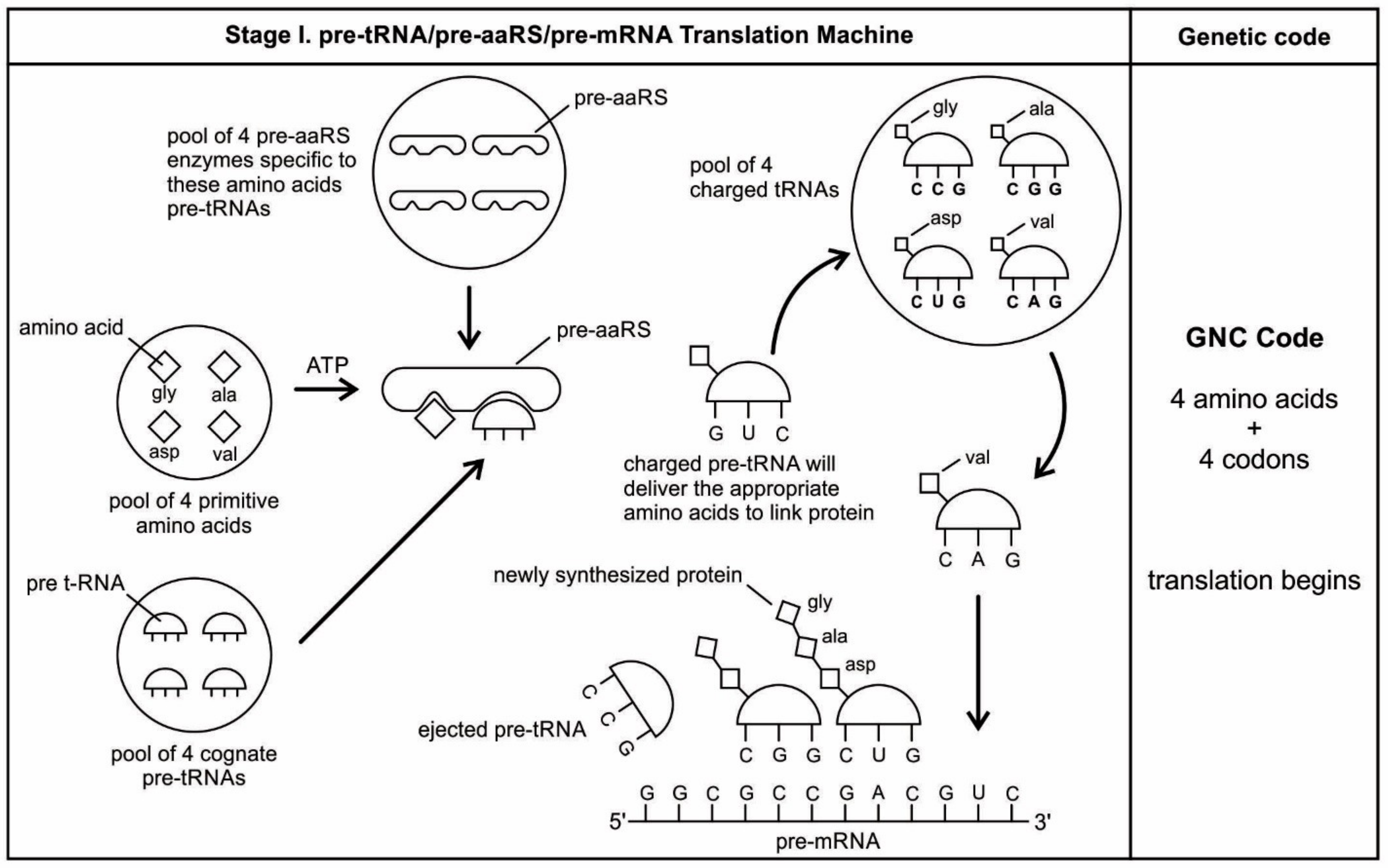
Here, we propose a new model for the synthesis of custom-made mRNA by tRNA. The evolution of non-random coding mRNA served as the first medium for genetic information that coincided with the development of the genetic code and protein synthesis. As the tRNA molecules began to recognize and react with certain amino acids, they need a separate storage device for safe keeping the information of amino acid assignment. Because the selection of mRNA exclusively depends on codon-anticodon interaction, tRNA begins to make a specific strand of mRNA for the storage of amino acid information (otherwise, it is difficult to see how else mRNA molecules could have become involved with coding the strings of amino acids in a specific manner). We suggest the origin of a new generation of ancestral mRNAs—pre-mRNAs, were created by pre-tRNAs step-by-step. These newly synthesized pre-mRNAs have direct preferences for the amino acids that they tend to encode.
In our model, pre-tRNA molecules begin to select codons via base pairing with their anticodons; these short codon segments are linked to create a longer strand of pre-mRNA step-by-step for storing genetic information. In the pre-tRNA molecule, the site of attachment of the appropriate amino acid is proximate to the anticodon, making the communication between two active sites easier (
Figure 5A,B). The physical proximity of the anticodon and the acceptor stem in ancestral pre-tRNA molecules is relevant to a long-sought goal-deriving amino acid/codon pairing rules from an ancestral nucleotide-based receptor-ligand recognition system [
66]. A crucial aspect of the origin of pre-mRNA is that codon units are not just randomly added. Instead, the anticodon of pre-tRNA acts as a template to select the matching codon of a pre-mRNA strand. Using the base pairing mechanism, each anticodon of a charged pre-tRNA molecule begins to attract corresponding nucleotides from the prebiotic pool by base pairing (
Figure 5D). After hybridization with anticodons, these triplet nucleotides begin to cluster and link together to form small chains of oligonucleotide with codon bases. Several small oligonucleotide chains begin to link to form a longer strand of a pre-mRNA molecule that becomes a database for storing the information of several amino acids (
Figure 5E). This coded pre-mRNA became the binding partners for pre-tRNA, enhancing mutual stability and instant cognition. This is a turning point in the origin of translation when a pre-mRNA molecule becomes a digital strip for the storage of genetic information in a separate device in the nucleotide language. Translation is easier to evolve, logically as well as chemically, if there is already a triplet-amino acid assignment that is present. Eventually, several strands of pre-mRNA are joined to form a longer strand of pre-mRNA. These pre-mRNA genes are very short, no longer than 30 to 80 nucleotides. The main feature of pre-mRNA is its heterogeneity for information content. A triplet code sequence with a random codon assignment has very high information content in protein synthesis. With different combinations of codons and varied lengths of pre-mRNA strand, a wide range of amino acid information could be stored for the synthesis of longer protein chain (
Figure 5F).
With the emergence of pre-mRNA, the information of anticodon assignment of large pre-tRNA populations can be transferred and stored in a codon message, along the strand of a pre-mRNA molecule. Along the linear strand of a pre-mRNA molecule, digital information for coding amino acids symbiotically emerged with the help of the anticodon of pre-tRNA molecules. Biological information was not only concentrated, but also specified along the strand of a pre-mRNA molecule. Charged pre-tRNA becomes the carrier of a specific amino acid that attached to the matching codon of pre-mRNA.
During the interaction of charged pre-tRNA with pre-mRNA, each aminoacyl pre-tRNA (aa-pre-tRNA) molecule transported and selected specific amino acids for protein synthesis. This is how information enters into the codon of the pre-mRNA molecule in a storage format for a specific amino acid via the anticodon. The information is laid down in the sequences of pre-mRNA, whose quantity is expressed by the lengths of those sequences. These base-pairing attachments between charged pre-tRNA and pre-mRNA provided the structural basis for translation.
The aa-pre-tRNA brings this specific amino acid to this pre-mRNA site during translation, where its anticodon binds to the complementary codon. Initially, four short oligonucleotides, each with a specific codon, were formed and joined in different combinations, specifying four amino acids, such as valine, alanine, aspartic acid, and glycine [
88]. This is the first stage of the origin of translation along with the genetic code, involving four amino acids, in which a small number of amino acids were coded by a small number of triplets (
Figure 5D). These four amino acids were readily available from the prebiotic vent environment. These oligonucleotides with codons are linked together by random combinations to form a pre-mRNA strand with a coded message (
Figure 5E). Once the base sequence of pre-mRNA is stored for a number of amino acids, a rudimentary translation begins to initiate between pre-tRNA and pre-mRNA to synthesize the protein products that provide some modest catalytic, structural, and binding features in the peptide/RNA world. Most likely, the code assignments and the translation mechanism evolved together [
75]. Pre-mRNA molecules, which were customized by pre-tRNA, multiplied in the vent environment and linked into longer strands of pre-mRNA to become a genetic reservoir, a digital recipe for proteins synthesis. However, at this stage, pre-mRNA can contain limited genetic information for four amino acids or their multiplied combinations.
During the initial translation process, each pre-tRNA carries its corresponding amino acid on its end (
Figure 5F). When a charged pre-tRNA recognizes and binds to its corresponding codon of pre-mRNA, then the growing amino acid chain transfers to the single amino acid of the pre-tRNA. The pre-tRNA molecule begins to translate the codon of the pre-mRNA molecule in the 5´ to 3´ direction. The codon for the first amino acid in the chain (the amino end of the protein) is always at the 5´-end of the pre-mRNA. Likewise, the codon for the last amino acid in the chain is at the 3´-end of the pre-mRNA.
As the translation began along the strand of pre-mRNA, the triplet GUC coded for the amino acid valine. An aminoacyl pre-tRNA entered the site, where it then hybridized the codon. Here, a ribozyme, the precursor to peptidyl transferase of ribosome, performed two critical functions. First, it detached the valine from its pre-tRNA, which was ready to make a growing amino acid chain and released the pre-tRNA. Second, it catalyzed the formation of a peptide bond between that amino acid and the one that was attached to the next codon site. The first pre-tRNA, carrying the amino acid glycine, paired with the codon GCC. With the arrival of the second pre-tRNA, carrying valine, the first pre-tRNA, like a runner in a relay race, passed its glycine to the next, linking with valine and it was ejected. The third pre-tRNA with anticodon CUC hybridized with the next codon, GAC, bearing the aspartic acid, and picked up the link of glycine and valine. The next step repeats when a new aminoacyl pre-tRNA prepares to attach to the next codon site CGG for alanine. Here, it would receive the newly formed polypeptide link of valine-glycine-aspartic acids. To this link, alanine would be added. This is the way that a string of bases of pre-mRNA is translated into a sequence of amino acids. The released amino acids chain of valine, glycine, aspartic acid, and alanine are joined together by a peptide bond to form a newly synthesized protein (
Figure 5F). Ribozymes functioned as a catalyst to break the acyl bond holding the growing amino acid chain on the pre-tRNA, and link the new incoming amino acid to the protein chain by a peptide bond. Those ribozymes that were involved in the protein synthesis were the precursors for the peptidyl transferase of the larger unit of the ribosomes.
The association between amino acids and codons—for example, between GUC and valine—is called the code. In this way, the genetic code begins to translate in a rudimentary form, as the short chain of proteins is built according to the instruction from the linear order of codons on the pre-mRNA. The process continues until the pre-tRNA molecule reaches the last codon in the pre-mRNA strand. It stops because there are no more codons to match. The ribozyme is clipped off by the completed protein chain. Once the complete protein is made, the pre-tRNA was discarded, and the pre-mRNA was broken down and its nucleotides recycled. The newly synthesized proteins functioned as enzymes for specific catalysis.
This initial code-programming and storage operation of the pre-mRNA by the pre-tRNA must have occurred within the protective environment of the protocells (
Figure 5C). By pairing with the anticodons of the pre-tRNAs, the codons of the pre-mRNA not only selected the appropriate amino acids, but they also help to immobilize the pre-tRNAs. To initiate primitive translation, the pre-mRNA strand needed a substrate where pre-tRNA molecules would sequentially bind one codon after another.
How the primitive translation machinery maintains its proper reading frame is a question of primary importance. In the absence of the ribosomes, the inner surface of the protocell membrane would have served as a substrate for holding the pre-mRNA in position for pairing with the anticodon (
Figure 5C). The spherical curved surface of the membrane probably facilitates the movement of pre-tRNA in downstream from 5´ to the 3´ ends of pre-mRNA during translation. This may be the beginning of the origin of reading frame, which is crucial for the reproducibility of translation; the codons of pre-mRNA should be read in a fixed direction with no gap between them.
The availability of several groups of new enzymes enlarged both the structural and the functional capabilities of the pre-mRNA and pre-tRNA molecules, evolving into the more efficient mRNA and tRNA. This evolutionary transformation was characterized by a progressive refinement of the translation system and an increase of the genetic code. As more and more pre-tRNA guided pre-mRNA molecules began to emerge, they were continuously replicated, increasing their population in the prebiotic pool, linking together in various combinations to form longer strands of mRNA molecules. tRNA and mRNA outnumber their precursors pre-tRNA and pre-mRNA through base pairing and replication. These longer mRNA genes arose as replication increased in accuracy. Each mRNA contained about 100 to 200 nucleotides (
Figure 5E).
5.8. The Origin of Ribosomes
Translation needs one more piece of the molecular machine to continuously make protein in an assembly line—the ribosome. Ribosomes link amino acids together in the order that is specified by mRNA molecules. They provide the environment for controlling the interaction between codons of mRNA and anticodons of aminoacyl-tRNA in the creation of proteins. The translation of encoded information of mRNA and the linking of amino acids that were selected by tRNAs are at the heart of the protein production process. Ribosomes can link amino acids together at a rate of 200/min. Therefore, small proteins can be made fairly quickly. Once a new protein chain is manufactured, the ribosome is released from protein synthesis to enter a pool of free ribosomes that are in equilibrium with separate small and large subunits [
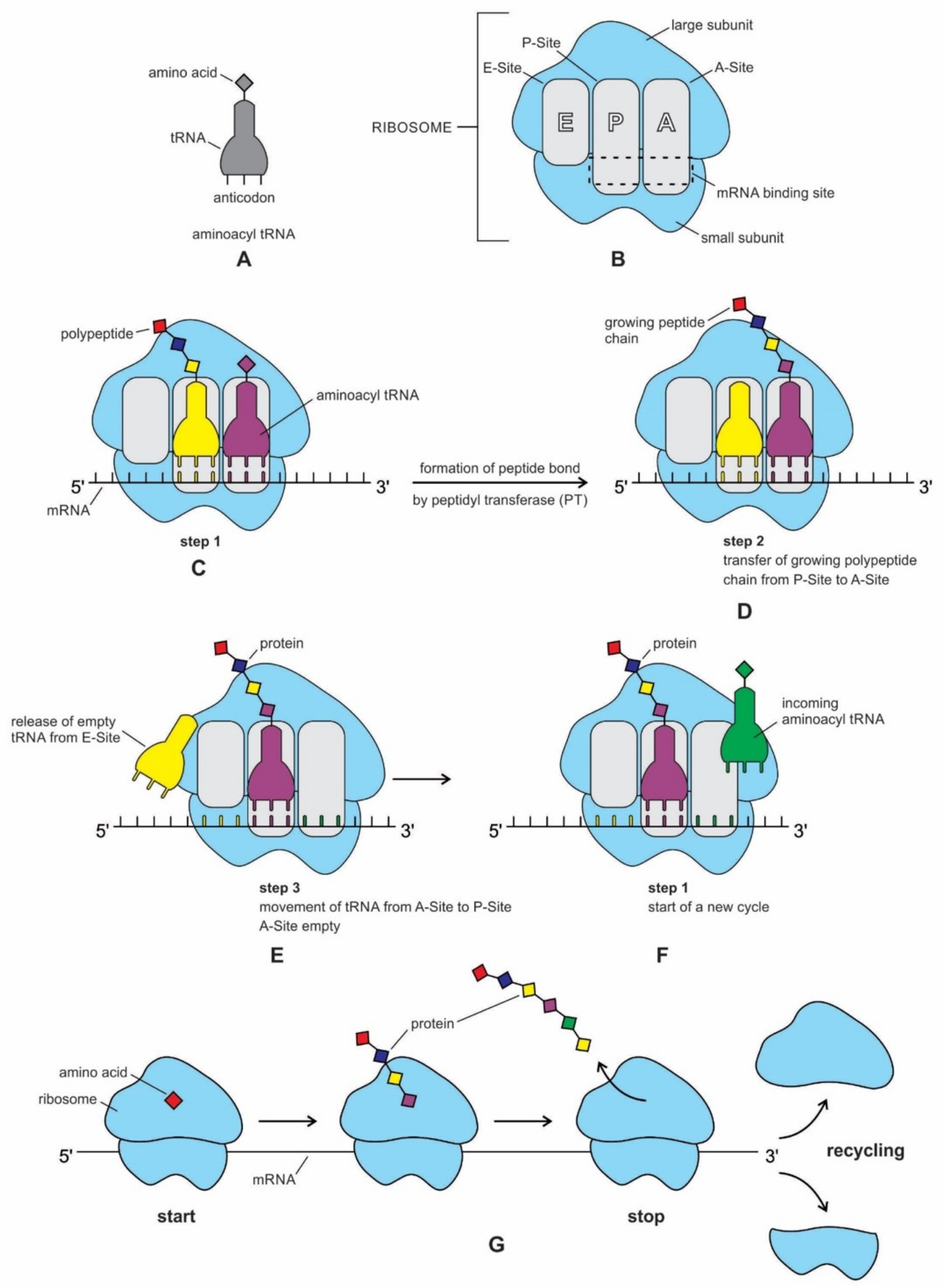
77].
The ribosome is composed of two-thirds of RNA and one-third protein. It is made of about 50 ribosomal proteins (r-protein) that are wrapped up with four ribosomal RNAs (rRNA) and it is therefore a ribonucleoprotein (
Figure 6). Although ribosomal proteins greatly outnumber ribosomal RNA, the rRNAs account for more than half the mass of the ribosome. A bacterial cell may contain as many as 20,000 ribosome complexes, which enable the continuous production of several thousand different proteins, both to replace degraded proteins and to make new ones for daughter cells during cell division. A ribosome physically moves along an mRNA strand, reads the codon sequences of mRNA, and catalyzes the assembly of amino acids into protein chains using the genetic code. It uses tRNAs to mediate the process of translation from the nucleotide language of mRNA into the amino acid language of proteins with the help of various accessory molecules. Each ribosome can bind one mRNA and up to three tRNAs. Central to the development of ribosomes are RNAs that spawn the tRNAs, and a symmetrical region that is deep within the large ribosomal RNA, where the peptidyl transferase reaction occurs [
77,
89,
90].
Recent bacterial ribosomes shed light on the origin, evolution, morphology, and composition of primitive ribosome that emerged in the peptide/RNA world. The bacteria have smaller ribosomes, termed 70S ribosomes, which are composed of two major subunits of unequal size, which are called the large (50S) and the small (30S) subunits; each consists of one or two RNA chains and scores of proteins (
Figure 6). The small subunit (SSU) is where mRNA and tRNA molecules interact to read the genetic code, and the large subunit (LSU) is where the growing protein chain is synthesized from the amino acids that are attached to tRNAs. Thus, the small subunit is mainly decoding mRNA, but the large subunit mainly has a catalytic function. In the large subunit, rRNA performs the function of an enzyme and it is termed as a ribozyme. In prokaryotic ribosomes, the small subunit, 30S, is made of one ribosomal RNA and 21 ribosomal proteins, while the large subunit, 50S, is made of two ribosomal RNAs and 31 ribosomal proteins. The two subunits fit snugly in a slot, through which a strand of the mRNA molecule runs between them, after the fashion of a tape through a cassette player. The ribosome glides through the mRNA tape, which then carries out its instructions bit by bit, linking the amino acids together, one by one in a specified sequence, until an entire protein has been synthesized. The ribosomal RNAs are programmed to recognize the codon as it appears on mRNA. When the production of a specific protein is finished, the two subunits of ribosome drift apart [
89,
90]. Ribosomes only have a temporary existence. The large and small subunits of the ribosome undergo a cycle of association and dissociation during each round of translation. Similarly, once the protein is made, mRNA is broken down and the nucleotides are recycled.
The ribosome evolved prior to the emergence of DNA and the cellular life in the peptide/RNA world. Ribosome evolution is intricately linked to the prior evolution of mRNA, tRNA, and primitive form of the genetic code and translation. The origins and evolution of ribosomes remain printed in the biochemistry of extant life and in the structure of the ribosome. Most theories propose that the ribosome was a functional takeover of a primitive RNA-based translation system in a coordinated series of chemical reactions. RNA is thought to be responsible for the bulk of the ribosome’s work. Recent structures of ribosomes have unambiguously shown that the essential functions of the ribosome, such as decoding, peptidyl transfer, and translocation, all appear to be mediated by RNA [
91]. Phylogeny of ribosome suggests that the origin of rRNA is linked to accretionary tRNA building blocks that gave rise to functional rRNA [
20]. The decoding center where mRNA is located in the small subunit and it is primarily formed from 16S rRNA. The rRNAs are folded into highly compact and precise three-dimensional structures that form the core of the ribosome. The rRNAs give the ribosome its overall shape. Thus, the widely popular concept of ‘the ribosome is a ribozyme’ was born; the ribozymes must have preceded coded protein synthesis [
39].
In recent times, the role of proteins in the origin of ribosomes is gaining currency, implying that the ribosome may have first originated in a peptide/RNA world, where both amino acids and a variety of enzymes were available [
9,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
92]. Ribosomal proteins are not passive contributors to ribosome function. They are generally located on the surface, where they fill the gaps and crevices of the folded rRNA. The main role of the ribosomal proteins seems to fold and stabilize the rRNA core, while permitting the changes in rRNA conformation that are necessary for this RNA to catalyze efficient protein synthesis. The ribosomal proteins provide the structural framework for the 23S rRNA, which actually carries out the peptidyl transferase reaction. In the absence of ribosomal proteins, 23S rRNA is unable to serve as a peptidyl transferase activity. The assembly of large and small subunits that are dependent upon ribosomal proteins [
17,
92]. Several ribosomal proteins assist in the assembly of the large subunit by providing unstructured, highly positively charged protein sequences that bind amino RNA segments together and extend to the center of the subunit [
92]. These extensions cooperatively fold with ribosomal proteins to produce the small subunit.
Why would an RNA structure evolve to make proteins if the protein did not already exist that would confer a selective advantage on the ribosomes capable of synthesizing them? The availability of even simple proteins could have significantly enlarged the otherwise limited catalytic function of RNA. Many prebiotic protein enzymes carried out several key functions in the primitive translation system. Moreover, the production of simple proteins had already commenced through the interactions of mRNA/tRNA/aaRS, before the origin of ribosome (
Figure 5). Perhaps ribosomal proteins were synthesized during the primitive translation system, which were then recruited to build the ribosome step-by-step. RNAs and proteins developed a symbiotic relationship to create ribosomes in the peptide/RNA world [
17,
18,
19]. These r-proteins took an active part in stabilizing the evolving ribosomes and in interacting with many rRNA sequences. Because the number of proteins greatly exceeded the number of RNA domains, it can hardly come as a surprise that every rRNA domain interacted with multiple proteins in ribosomes [
91]. Ribosomes are not entirely ribozymes, but are more accurately ribonucleoprotein (RNP), a complex that can have as many as 62 r-proteins, with only three rRNA molecules (
Figure 6). Virtually all r-proteins are in contact with the rRNA. Accordingly, it makes sense that this assemblage is a result of a long and complicated process of gradual coevolution of rRNAs and r-proteins. Both the assembly and synthesis of the ribosomal components must occur in a highly coordinated fashion [
20]. Their phylogenetic analysis reveals that the ribosomal protein/rRNA coevolution manifested throughout the prebiotic synthesis process, but the oldest protein (S12, S17, S9, L3) appeared together with the oldest rRNA substructures that were responsible for both the decoding and ribosomal dynamics 3.3-3.4 Ga. Although protein synthesis is largely carried out by different kinds of RNA molecules within the ribosome, such as mRNA, tRNA, rRNA, and peptidyl transferase, aminoacyl synthetase (aaRS) played a crucial role as a protein enzyme that attached the appropriate amino acid onto its tRNA during protein synthesis. The synthetase, in terms of importance, is equal to the tRNAs in the decoding process, because it is the combined action of synthetases and tRNAs that allows each codon in the mRNA molecule to associate with its proper amino acid. Similarly, both rRNA and the 50S subunit proteins are necessary for the peptidyl transferase activity during peptide bond formation, but the actual act of catalysis is a property of the ribosomal RNA of the larger subunit (
Figure 6). The cumulative conclusion that seems to be most in accord with biochemical evidence is that the peptide/RNA world preceded ribosome.
The accretion model describes the origin and evolution of ribosomes [
20]. Given that the ribosome is quite ancient, it is likely that rRNAs and r-proteins coevolved to build this complex nanomachine. Ribosomes, like the rings of a tree, contain the record of their history, spanning four billion years. Like rings in the trunk of a tree, the ribosome contains components that functioned on in its early history. It accreted to grow bigger and bigger over time. However, the older parts froze after they accreted, like the rings of a tree (
Figure 6). Recent phylogenetic work on ribosomal history suggests that both RNAs and proteins contributed to the formation of the ribosome core through accretion, recursively adding expanding segments [
20,
21]. Ribosomes contains life’s most ancient and abundant polymers, the oldest fragments of RNA and protein molecules. It most likely a molecular relic of the peptide/RNA world [
9].
Both ribosomal subunits have separate functions. Peptide bond formation occurs at the peptidyl transferase center (PTC) of the large subunit, whereas the mRNA sequences are decoded on the small subunit. mRNA decoding contributes to the specificity of protein synthesis on the ribosome. In isolation, both of the subunits can perform their respective functions (
Figure 6). By itself, the large subunit will catalyze the formation of peptide bonds between aminoacyl-tRNA-like substrates. By itself, the small subunit binds mRNA, and when mRNA is bound, it will bind tRNAs in a codon-specific manner. In an RNA world scenario, the ribosome originated in the peptidyl transferase center of the large ribosomal subunit [
93,
94]. There are no r-proteins that are close to the reaction site for protein synthesis. This suggests that the protein components of the ribosome do not directly participate in the peptide bond formation catalysis, but rather the proteins act as a scaffold that may enhance the ability of rRNA to synthesize protein. Ribosomes themselves, although being fundamentally ribozymes in nature, still require r-proteins to fold their rRNAs into biologically active conformations and to optimize the speed and accuracy of their functions [
85]. The ribosomal surface is an integrated patchwork of rRNAs and r-proteins.
Currently, there is a debate regarding the origin of the ribosomal subunits: which unit came first, the small or the large subunit? It is likely that the PTC of large ribosomal subunit evolved from pre-tRNA molecules by duplication of the minihelix [
81]. In this view, the simple function of peptide bond formation at the PTC site came first, and the specifications that were based on the codon sequence came later. In other words, the large subunit of the ribosome came first, followed by the addition of the small unit. However, these proposals do not link the protein synthesis to RNA recognition and do not use a phylogenetic comparative framework to study ribosomal evolution.
Other authors who favor the small unit of ribosome as the first, deduced from the phylogeny of ribosome, offer a contrasting view of the origin of ribosomal subunits [
20]. The study suggests that the components of the small ribosomal subunit evolved earlier than the catalytic peptidyl transferase center of the large ribosomal subunit. In this view, the ribosomal RNA and proteins coevolved tightly, starting with the oldest proteins (S12 and S17) and the oldest rRNA helix in the small subunit (the ribosomal ratchet responsible for ribosomal dynamics), ending with the modern multi-subunit ribosome. A major transition in the evolution of ribosomes at around 4 Ga brought independently evolving subunits together by infolding the inter-subunit contacts and interaction with full cloverleaf tRNA structures.
In our view, both the small subunit and the large subunit of the ribosome simultaneously appeared and worked together, because the decoding of mRNA and the peptide bond formation were both essential components during protein synthesis. These two subunits might have coevolved to join during translation and separate after protein synthesis. The rRNAs are folded into highly compact, precise three-dimensional structures to form the core of the ribosome, whereas the r-proteins are generally located on the surface, where they fill the gaps and crevices of the folded RNA and act to fold and stabilize the core [
95]. As these two subunits expanded through accretion, eventually arriving at the size of the bacterial ribosome, the accretion stopped, they then bound together during protein synthesis, and finally spilt apart when the ribosome finished reading its mRNA molecule (
Figure 6).
If the fundamental functions of the ribosome are based on rRNA, then why are there so many ribosomal proteins, some of which are highly conserved? One explanation is the rRNA does not fold into its functional state in the absence of r-proteins. Another reason for the presence of proteins in ribosomes is that they improve the efficiency and accuracy of the translation [
93]. Both rRNAs and r-proteins work cooperatively in ribosomes to perform the multitask procedure of protein synthesis. Harish and Caetano-Anolles suggested that functionally important and conserved regions of the ribosome were recruited and could be relics of an ancient peptide/RNA world [
20]. The corollary is that a fully functional biosynthetic mechanism that is responsible for primordial peptides and ancient r-proteins must have existed that in time was superseded by the ribosome.
According to this accretionary model, very early in ribosomal evolution, rRNA helices interacted with r-proteins to progressively form a core that mediated nucleotide interactions, which later served as the center for the coordinated and balanced RNP (ribonucleoprotein) accretion that evolved into our modern ribosomal function [
20]. The early existence of smaller functional units of ribosome, which are capable of carrying out different translational steps, such as peptidyl transferase, decoding, and aminoacylation, along with the development of A, P, and E sites for the positioning of tRNA molecules, can be inferred from the phylogeny. These small functional RNA/protein units were incrementally accreted and then refined by the incorporation of additional rRNA and r-protein molecules. Similarly, the first atomic resolution of the larger of the two subunits of the ribosome suggests that the RNA components of the large subunit accomplish the key peptidyl transferase reaction [
96]. Thus, rRNA does not exist as the framework to organize catalytic proteins. Instead, the proteins are the structural units and they help to organize the key ribozyme. A ‘pure’ RNA world is incompatible with the existence of the coevolutionary pattern that is proposed for ribosomal molecules.
Perhaps rRNA, such as noncoding ribozymes, acquired amino acids as cofactors, making them more efficient catalysts. By using cofactors, the range and specificity of catalytic activity can be increased. Ribozymes would have been in greater need of cofactors than protein enzymes, because, without them, the range of reactions that they can catalyze is much smaller [
46].
In our endosymbiotic model, rRNAs and r-proteins were brought into close proximity within the plasma membrane to form the building block of the primordial ribosome. The origin of the ribosome precursor through fusion and the accretion of the key components of these ribosomal RNA and protein molecules is the likely scenario. The rRNA and r-protein molecules began to fuse because of a chiral preference and then formed the rudimentary ribosomes. Once the core of the ribosome formed, the mRNA and tRNA molecules were recruited to help in translation through a trial and error method. Once a true mRNA and the core small subunit of ribosome were in place, the ribosome would become increasingly complex by adding early conserved rRNA and r-proteins. Ribosomal proteins played an important role in supporting the ribosome structure and in promoting translation. With the onset of operational coding, tRNA began to assemble amino acids into long chains of proteins. Here, we suggest that a ribosome-like entity was one of the key intermediates between prebiotic and cellular evolution, which formed by endosymbiosis and the fusion of rRNA and r-protein molecules. Once ribosomes were installed inside the protocell membranes, the translation system was greatly improved.
In vitro constructions of ribosomes can shed new light on the mechanism of protein synthesis and provide deeper insights into the way that nature has assembled this complex machine. Working with
E. coli cells, natural ribosomal proteins were combined with synthetically made rRNA, which self-assembled in vitro to create semi-synthetic, functional ribosomes [
96,
97,
98]. Comprising 57 parts—three strands of rRNAs and 54 proteins—an artificial ribosome (termed Ribo-T), in which two subunits are tethered together by a short length of RNA, is able to carry out normal translation and pump out custom-made proteins. The ability to make ribosomes in vitro is a process that mimics nature and opens up new avenues for the study of ribosome synthesis, suggesting the coevolution of ribosomal RNAs and proteins.
5.9. Protein Synthesis
We have now reviewed the emergence of all major components of the translation machinery for protein synthesis. Translation of the mRNA template converts nucleotide-based genetic information into the ‘language’ of amino acids to create a protein product. Translation requires the input of an mRNA template, tRNAs, aminoacyl-tRNA synthetases, ribosomes, and various enzymatic factors. The tRNAs function as the adaptor molecules that transport amino acids to ribosomes in response to codons in mRNAs, where peptidyl transferase catalyzes the addition of amino acid residues to the growing protein chain in protein synthesis by means of peptide bonds. The ribosomes serve as the sites for protein synthesis and they link amino acids together in the order specified by mRNA. They always translate the mRNA from the 5’ to the 3’ direction, like a sliding machine.
Proteins have a modular chemical structure that allows for the construction of widely different molecular machines using the same basic set of amino acids, each with a different size and chemical character. Protein synthesis requires the concerted effort of dozens of different enzymes. 20 tRNA molecules, each with their own dedicated synthetase enzyme, are built for 20 amino acids. Modern protein synthesis proceeds with the participation of 20 amino acids, tRNA, mRNA, ribosomes, various enzymes, including aminoacyl-tRNA synthetase, ribozymes, peptidyl transferase, and a considerable number of proteinous factors, ATP, GTP, etc. More than 120 species of RNAs and proteins are involved in the process of protein synthesis [
65]. These biomolecules were related, encapsulated, and interacted with each other in complex ways, like an autopoietic machine. Yet, the whole series of molecules in the translation process functioned with astounding precision, in a kind of molecular choreography, which gave birth to the universal genetic code.
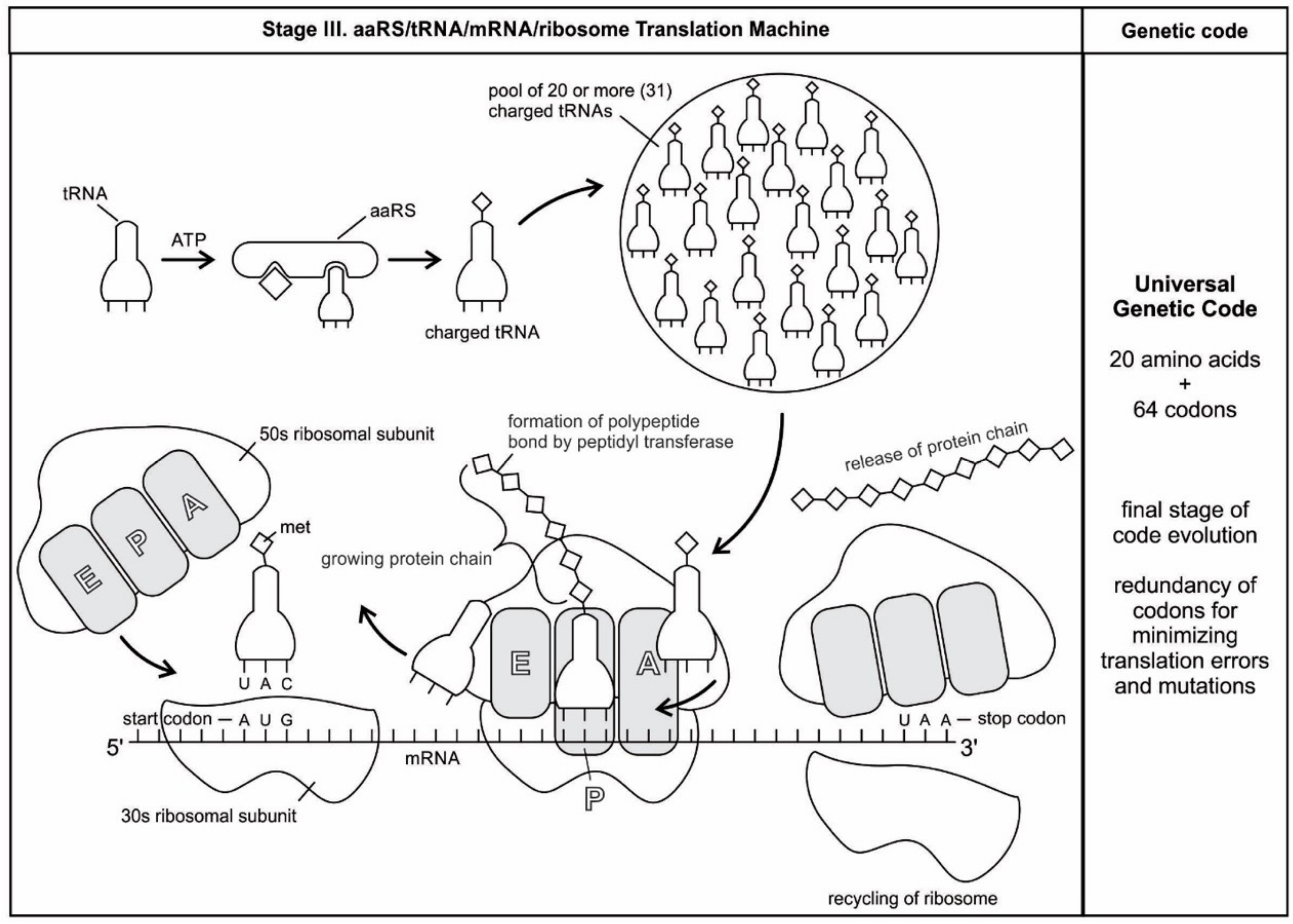
The structure and function of the modern ribosome during translation are well-known in the literature and they will not be repeated here [
77,
99]. In the ribosome, there are three stages and three operational sites that are involved in the protein production line and all work in harmony. During the initiation stage, a small ribosome subunit links onto the ‘start end’ of an mRNA strand. Aminoacyl-tRNA also enters site A of the ribosome. The production of the protein has now been initiated. The second stage, elongation, consists of joining amino acids to the growing protein chain, according to the sequence that was specified by the message. The incorporation of each amino acid occurs by the same mechanism. In the termination stage, the ribosome reaches the end of the mRNA strand, a terminal, or ‘end of the protein code’ message. This registers the end of production for the particular protein that was coded by this strand of mRNA (
Figure 7).
Translation is not the end of the protein synthesis process. Once released from the ribosome, the long chain of amino acids will spontaneously fold in intricate contortions into a unique three-dimensional configuration and proper characteristic shape: some parts form sheets, while others stack, curl, and twist into spirals. The sequence of amino acids determines the shape and conformation of a protein and, thereby, all of its physical and chemical properties. A protein molecule spontaneously folds during or after biosynthesis, but the folding process depends on the solvent, the concentration of salts, the temperature, the possible presence of cofactors, and the molecular chaperons [
99]. Proteins must fold in specific ways to function properly.