Figure 1.
Molecular cellular pathology of SARS-CoV-2 infection: Panel (
A) Schematic Illustration of SARS-CoV-2 viral RNA genome in relation to order of expression and functional incorporation with the Virion particle. The genome comprises a 5′-untranslated region (5′-UTR), open-reading frames (ORFs) 1a and 1b encoding nonstructural proteins 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), helicase (Hel), and RNA-dependent RNA polymerase (RdRp) as well as accessory proteins, and the structural S protein (S), E protein (E), M protein (M), and Nucleocapsid protein (N) in ORFs 2,4,5 and 9. Panel (
B). Schematic of virion fusion mediated by Spike protein attachment to the ACE2 receptor and accessory cleavage of the Spike protein quaternary complex, resulting in fusion peptide exposure and binding to target cell plasma membrane. Focal, host cell and viral envelope lipid bilayer membrane fusion and viral RNA ingress of the target host cell. Shedding of Nucleocapsid and early ORF expression to take control of the cell and inactivate internal anti-viral proteins (e.g., Ubiquitin and RNase). Late ORF expression: replication of viral RNA genome and expression of, via post-translational processing mechanisms of the endoplasmic reticulum and Golgi apparatus (e.g., glycosylation), membrane embedded proteins that are responsible for infectivity including the Spike protein complex. Copies of SARS-CoV-2 vRNA are packaged into lipid bilayer membrane envelopes containing multiple copies of S, E, M and N, glyco- and phospho- proteins. Inclusion transport vesicle, packed with enveloped virions, fuses with host cell membrane releasing infective SARS-CoV-2 virion. Panel (
C). Electro micrographs of multiple virions attacking the cell membrane (
i), viral genome expression subsuming all functions of the cell to form thousands of virion copies within inclusion vesicles (
ii) and finally release of multiple virions by fusion of inclusion packaging vesicles with the cell surface plasma membrane of the infected host cell (
iii). Original composite figure adapted in parts from Depfenhart et al. [
7] Kupferschmidt and Cohen [
8] and Mason [
9]. Scale bar: 1000 nm
Figure 1.
Molecular cellular pathology of SARS-CoV-2 infection: Panel (
A) Schematic Illustration of SARS-CoV-2 viral RNA genome in relation to order of expression and functional incorporation with the Virion particle. The genome comprises a 5′-untranslated region (5′-UTR), open-reading frames (ORFs) 1a and 1b encoding nonstructural proteins 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), helicase (Hel), and RNA-dependent RNA polymerase (RdRp) as well as accessory proteins, and the structural S protein (S), E protein (E), M protein (M), and Nucleocapsid protein (N) in ORFs 2,4,5 and 9. Panel (
B). Schematic of virion fusion mediated by Spike protein attachment to the ACE2 receptor and accessory cleavage of the Spike protein quaternary complex, resulting in fusion peptide exposure and binding to target cell plasma membrane. Focal, host cell and viral envelope lipid bilayer membrane fusion and viral RNA ingress of the target host cell. Shedding of Nucleocapsid and early ORF expression to take control of the cell and inactivate internal anti-viral proteins (e.g., Ubiquitin and RNase). Late ORF expression: replication of viral RNA genome and expression of, via post-translational processing mechanisms of the endoplasmic reticulum and Golgi apparatus (e.g., glycosylation), membrane embedded proteins that are responsible for infectivity including the Spike protein complex. Copies of SARS-CoV-2 vRNA are packaged into lipid bilayer membrane envelopes containing multiple copies of S, E, M and N, glyco- and phospho- proteins. Inclusion transport vesicle, packed with enveloped virions, fuses with host cell membrane releasing infective SARS-CoV-2 virion. Panel (
C). Electro micrographs of multiple virions attacking the cell membrane (
i), viral genome expression subsuming all functions of the cell to form thousands of virion copies within inclusion vesicles (
ii) and finally release of multiple virions by fusion of inclusion packaging vesicles with the cell surface plasma membrane of the infected host cell (
iii). Original composite figure adapted in parts from Depfenhart et al. [
7] Kupferschmidt and Cohen [
8] and Mason [
9]. Scale bar: 1000 nm
![Diagnostics 10 00746 g001]()
Figure 2.
Process of virion enrichment within a biological sample and extraction/solubilisation of Spike (S) and other viral envelope protein for MALDI ToF MS analysis. Panel (A): Biological samples containing virus are mixed 1 to 1 v/v with ice cold acetone and centrifuged at 16,000× g for 30 min at 4 °C. The supernatant containing smaller non-precipitated solutes is discarded. The pellet is enriched with viral particles and kept for analysis. Acetone treatment inactivates envelope virus by deformation and partial fragmentation of the viral envelope and embedded protein Structures A. Panel (B): The pellet is resuspended in 10 to 100 uL of MALDI -ToF Mass spectrometry compatible dissolution and solubilisation buffer. Termed LBSD-X this buffer does not suppress ionisation and contains a detergent at a concentration optimised to release viral envelope embedded proteins together with non-embedded viral proteins. It also contains Dithiothreitol (DTT) in order to further reduce disulphide bonds so that quaternary and tertial structures are fully disrupted and monomers, polypeptide chains and glyco-polypeptides are liberated for detailed mass analysis.
Figure 2.
Process of virion enrichment within a biological sample and extraction/solubilisation of Spike (S) and other viral envelope protein for MALDI ToF MS analysis. Panel (A): Biological samples containing virus are mixed 1 to 1 v/v with ice cold acetone and centrifuged at 16,000× g for 30 min at 4 °C. The supernatant containing smaller non-precipitated solutes is discarded. The pellet is enriched with viral particles and kept for analysis. Acetone treatment inactivates envelope virus by deformation and partial fragmentation of the viral envelope and embedded protein Structures A. Panel (B): The pellet is resuspended in 10 to 100 uL of MALDI -ToF Mass spectrometry compatible dissolution and solubilisation buffer. Termed LBSD-X this buffer does not suppress ionisation and contains a detergent at a concentration optimised to release viral envelope embedded proteins together with non-embedded viral proteins. It also contains Dithiothreitol (DTT) in order to further reduce disulphide bonds so that quaternary and tertial structures are fully disrupted and monomers, polypeptide chains and glyco-polypeptides are liberated for detailed mass analysis.
![Diagnostics 10 00746 g002]()
Figure 3.
Mass spectral profiles of Pseudo Virus expressing SARS-CoV-2 Spike S protein grown in culture, SARS-CoV-2 virus grown in vitro and mass spectra of gargle/saliva spiked with culture media from cells infected with SARS-CoV-2: S proteolytic fragments S1 and S2 were seen in all preparations and S2b only in serum free samples. Viral envelope proteins (VEPs) became more prevalent in live virus culture and Ig light chains and IgA heavy chain were additional peaks found in gargle/saliva samples. HEK293T/17 cells were seeded into 6 well cell-culture plates and co-transfected after 24 h with p8.91 (gag-pol expression plasmid), pCSFLW (luciferase reporter plasmid), and a viral glycoprotein expression plasmid encoding the spike protein of either SARS-CoV-2. The transfection mixture was prepared in OptiMEM using FuGENE-HD as a transfection reagent; cell culture media was replaced prior to transfection. The cell supernatant was collected via syringe 48 h after transfection and filtered through a 0.45 µm filter to harvest the lentiviral pseudo-type particles [
28]. Pseudo virus expressing post translationally modified and conformational function SARS S protein and live SARS-CoV-2 virus where grown in culture, under category 3 containment conditions, on Vero-Green Monkey cells. Virions present in the culture media were filtered (0.45 µm), acetone precipitated-enriched as described and all viral envelope proteins, including S protein fragments, extracted and solubilized using extraction formulation LBSD-X buffer and detected as mass peaks by MALDI ToF MS. Scale bar: 1000 nm
Figure 3.
Mass spectral profiles of Pseudo Virus expressing SARS-CoV-2 Spike S protein grown in culture, SARS-CoV-2 virus grown in vitro and mass spectra of gargle/saliva spiked with culture media from cells infected with SARS-CoV-2: S proteolytic fragments S1 and S2 were seen in all preparations and S2b only in serum free samples. Viral envelope proteins (VEPs) became more prevalent in live virus culture and Ig light chains and IgA heavy chain were additional peaks found in gargle/saliva samples. HEK293T/17 cells were seeded into 6 well cell-culture plates and co-transfected after 24 h with p8.91 (gag-pol expression plasmid), pCSFLW (luciferase reporter plasmid), and a viral glycoprotein expression plasmid encoding the spike protein of either SARS-CoV-2. The transfection mixture was prepared in OptiMEM using FuGENE-HD as a transfection reagent; cell culture media was replaced prior to transfection. The cell supernatant was collected via syringe 48 h after transfection and filtered through a 0.45 µm filter to harvest the lentiviral pseudo-type particles [
28]. Pseudo virus expressing post translationally modified and conformational function SARS S protein and live SARS-CoV-2 virus where grown in culture, under category 3 containment conditions, on Vero-Green Monkey cells. Virions present in the culture media were filtered (0.45 µm), acetone precipitated-enriched as described and all viral envelope proteins, including S protein fragments, extracted and solubilized using extraction formulation LBSD-X buffer and detected as mass peaks by MALDI ToF MS. Scale bar: 1000 nm
![Diagnostics 10 00746 g003]()
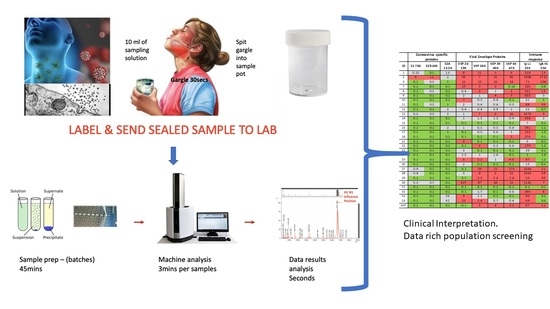
Figure 4.
Clinical diagnostic protocol of gargle saliva collection, laboratory processing and spectral data features: Panel (A) illustrates patient (home) sampling gargling 10 mL of water and spitting that into a sample pot. Panel (B) illustrates rapid processing where received gargle sample is filtered through 1.2 and 0.45 mm filters, 5 mL is recovered and is acetone precipitated. The pellet is resuspended in viral envelope dissolution and protein solubilization buffer before being applied to MALDI ToF plates and analyzed (3 min per sample); output data being processed by appropriate software. Panel (C) illustrates a reference spectra of SARS-CoV2 grown in vitro with spectral pattern of viral envelope proteins (VEPs) and fragments of coronavirus spike protein labelled. Panel (D) illustrates overall spectra of 5 unscreened volunteer samples overlaid. The spectra from subject ID2 is indicated (blue line). Regions corresponding to mass ranges for VEPs and large characteristic fragments of the Spike protein are illustrated. These are illustrated in juxta position to gargle/salivary Immunoglobulin light chain and Ig A heavy chain peaks. Also to note that S2a fragment is surrounded by a host of high intensity/abundance proteins, the origins of which may be a mix of other co-enriched (bacterial) micro-organism from the oral cavity. An additional high molecular mass glyco-protein type peak was also seen at with a maxima at 112,000 m/z (marked **) was only seen in this sample.
Figure 4.
Clinical diagnostic protocol of gargle saliva collection, laboratory processing and spectral data features: Panel (A) illustrates patient (home) sampling gargling 10 mL of water and spitting that into a sample pot. Panel (B) illustrates rapid processing where received gargle sample is filtered through 1.2 and 0.45 mm filters, 5 mL is recovered and is acetone precipitated. The pellet is resuspended in viral envelope dissolution and protein solubilization buffer before being applied to MALDI ToF plates and analyzed (3 min per sample); output data being processed by appropriate software. Panel (C) illustrates a reference spectra of SARS-CoV2 grown in vitro with spectral pattern of viral envelope proteins (VEPs) and fragments of coronavirus spike protein labelled. Panel (D) illustrates overall spectra of 5 unscreened volunteer samples overlaid. The spectra from subject ID2 is indicated (blue line). Regions corresponding to mass ranges for VEPs and large characteristic fragments of the Spike protein are illustrated. These are illustrated in juxta position to gargle/salivary Immunoglobulin light chain and Ig A heavy chain peaks. Also to note that S2a fragment is surrounded by a host of high intensity/abundance proteins, the origins of which may be a mix of other co-enriched (bacterial) micro-organism from the oral cavity. An additional high molecular mass glyco-protein type peak was also seen at with a maxima at 112,000 m/z (marked **) was only seen in this sample.
![Diagnostics 10 00746 g004]()
Figure 5.
Levels of proteolytic S protein fragments and viral-associated protein peaks in samples groups. Culture supernatant from Vero cells infected with Sars-Cov-2 were spiked into ten gargle/saliva samples at 1–10%. The viral load in these culture media was determined as at 104 to 105 PFU/mL. Gargle/saliva were collected from two individuals, with and without a cough, who were confirmed as RT-PCR Covid-19 negative at the time of gargle/saliva collection. These were analysed 20 times each. A series of 30 gargle/saliva was collected from volunteers, but COVID-19 PCR status at time of sampling was not known. These were analysed only once each. All samples were acetone precipitated, subjected to viral envelope protein solubilisation and analysed by MALDI-ToF mass spectrometry. Target peak intensities were then measured. Panel (A) illustrates the levels of each analysed sample for Spike protein fragment S1 peak at 79,000 m/z; panel (B) Spike protein fragment S2b at 63,000 m/z and panel (C) Spike protein fragment S2a at 13,500 m/z. The maximal intensity was also measured for broad glycoproteins peaks, considered to originate from other viral envelope proteins (VEPS), at 26,000–28,000 m/z (vep26–28K–panel (D)), 34,000 m/z (vep34K panel (E)), 39,000-40,000 m/z (vep39–40K–panel (F)) and 45,000-47,000 m/z (vep45K–panel (G)). Red dots represent levels of peak intensity for the spiked gargle/saliva samples, orange dots represents levels of peak intensity for COVID-19 RT-PCR negative control 1 samples, grey dots represent levels of peak intensity for COVID-19 PCR negative control 2 samples and blue dots represent levels of peak intensity for unscreened volunteer gargle/saliva samples. Blue dots with red circle borders are the values recorded for volunteer subject ID2.
Figure 5.
Levels of proteolytic S protein fragments and viral-associated protein peaks in samples groups. Culture supernatant from Vero cells infected with Sars-Cov-2 were spiked into ten gargle/saliva samples at 1–10%. The viral load in these culture media was determined as at 104 to 105 PFU/mL. Gargle/saliva were collected from two individuals, with and without a cough, who were confirmed as RT-PCR Covid-19 negative at the time of gargle/saliva collection. These were analysed 20 times each. A series of 30 gargle/saliva was collected from volunteers, but COVID-19 PCR status at time of sampling was not known. These were analysed only once each. All samples were acetone precipitated, subjected to viral envelope protein solubilisation and analysed by MALDI-ToF mass spectrometry. Target peak intensities were then measured. Panel (A) illustrates the levels of each analysed sample for Spike protein fragment S1 peak at 79,000 m/z; panel (B) Spike protein fragment S2b at 63,000 m/z and panel (C) Spike protein fragment S2a at 13,500 m/z. The maximal intensity was also measured for broad glycoproteins peaks, considered to originate from other viral envelope proteins (VEPS), at 26,000–28,000 m/z (vep26–28K–panel (D)), 34,000 m/z (vep34K panel (E)), 39,000-40,000 m/z (vep39–40K–panel (F)) and 45,000-47,000 m/z (vep45K–panel (G)). Red dots represent levels of peak intensity for the spiked gargle/saliva samples, orange dots represents levels of peak intensity for COVID-19 RT-PCR negative control 1 samples, grey dots represent levels of peak intensity for COVID-19 PCR negative control 2 samples and blue dots represent levels of peak intensity for unscreened volunteer gargle/saliva samples. Blue dots with red circle borders are the values recorded for volunteer subject ID2.
![Diagnostics 10 00746 g005]()
Figure 6.
Gargle/saliva Immunoglobulin protein levels. Panel (A) are plots comparing the total Ig light chain and total IgA heavy chain peak intensity levels for two COVID-19 PCR negatives control individuals (grey and orange dots respectively) repeated 20 times each (n = 40), versus the same peaks intensity level of 30 volunteers (blue dots) who gave gargle/saliva samples but whose COVID-19 PCR status was unknown at the time. Blue dots bound by a red circle are the levels of total Ig light chains and IgA heavy chains found in volunteer ID2. Panel (B) are scatter plots of total Ig light chains (upper plot) and IgA heavy chain peak intensities (lower plot) versus VEPs peak intensities for the 30 volunteer gargle/saliva samples but whose COVID-19 PCR status was unknown at the time. No VEP level correlated with total Ig light chain level. Correlation were seen with Ig A heavy chain levels for vep39–40K (grey dots r2 = 0.60), vep44–47K (gold dots r2 = 0.53) and vep34k (orange dots r2 = 0.53) but not for vep26–28k (blue dots r2 = 0.06)
Figure 6.
Gargle/saliva Immunoglobulin protein levels. Panel (A) are plots comparing the total Ig light chain and total IgA heavy chain peak intensity levels for two COVID-19 PCR negatives control individuals (grey and orange dots respectively) repeated 20 times each (n = 40), versus the same peaks intensity level of 30 volunteers (blue dots) who gave gargle/saliva samples but whose COVID-19 PCR status was unknown at the time. Blue dots bound by a red circle are the levels of total Ig light chains and IgA heavy chains found in volunteer ID2. Panel (B) are scatter plots of total Ig light chains (upper plot) and IgA heavy chain peak intensities (lower plot) versus VEPs peak intensities for the 30 volunteer gargle/saliva samples but whose COVID-19 PCR status was unknown at the time. No VEP level correlated with total Ig light chain level. Correlation were seen with Ig A heavy chain levels for vep39–40K (grey dots r2 = 0.60), vep44–47K (gold dots r2 = 0.53) and vep34k (orange dots r2 = 0.53) but not for vep26–28k (blue dots r2 = 0.06)
![Diagnostics 10 00746 g006]()
Figure 7.
Illustration of clinical data output. Format is a heat map scoring of peak intensity relative to those values for SARS-2 coronavirus spike protein fragments and viral envelope proteins, measured in gargle/saliva containing 1–10% culture media of SARS-CoV-2 virus at 104 per ml PFU. Red indicating elevated, Grey indicating slight to moderate increase and Green indicating within normal range. Heat map associated with immunoglobulin intensities are based on the range found in the volunteers for which volunteer ID2 was identified as recovering from suspected COVID-19 infection. ID2 was repeated after 14 days and his scores are shown under ID44 *.
Figure 7.
Illustration of clinical data output. Format is a heat map scoring of peak intensity relative to those values for SARS-2 coronavirus spike protein fragments and viral envelope proteins, measured in gargle/saliva containing 1–10% culture media of SARS-CoV-2 virus at 104 per ml PFU. Red indicating elevated, Grey indicating slight to moderate increase and Green indicating within normal range. Heat map associated with immunoglobulin intensities are based on the range found in the volunteers for which volunteer ID2 was identified as recovering from suspected COVID-19 infection. ID2 was repeated after 14 days and his scores are shown under ID44 *.