Abstract
This study aims to investigate and assess salivary biomarkers and microbial profiles as a means of diagnosing periodontitis. A total of 121 subjects were included: 28 periodontally healthy subjects, 24 with Stage I periodontitis, 24 with Stage II, 23 with Stage III, and 22 with Stage IV. Salivary proteins (including active matrix metalloproteinase-8 (MMP-8), pro-MMP-8, total MMP-8, C-reactive protein, secretory immunoglobulin A) and planktonic bacteria (including Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Fusobacterium nucleatum, Prevotella intermedia, Porphyromonas nigrescens, Parvimonas micra, Campylobacter rectus, Eubacterium nodatum, Eikenella corrodens, Streptococcus mutans, Staphylococcus aureus, Enterococcus faecalis, and Actinomyces viscosus) were measured from salivary samples. The performance of the diagnostic models was assessed by receiver operating characteristics (ROCs) and area under the ROC curve (AUC) analysis. The diagnostic models were constructed based on the subjects’ proteins and/or microbial profiles, resulting in two potential diagnosis models that achieved better diagnostic powers, with an AUC value > 0.750 for the diagnosis of Stages II, III, and IV periodontitis (Model PA-I; AUC: 0.796, sensitivity: 0.754, specificity: 0.712) and for the diagnosis of Stages III and IV periodontitis (Model PA-II; AUC: 0.796, sensitivity: 0.756, specificity: 0.868). This study can contribute to screening for periodontitis based on salivary biomarkers.
1. Introduction
The diagnosis of periodontitis is conventionally based on clinical evaluation, including probing pocket depth, bleeding on probing, clinical attachment level, periodontal index and gingival index, and radiographic examinations. However, the conventional method has a limitation in that some parameters reflect only past evidence of inflammatory changes and do not show that this inflammatory change would progress or regress in the future [1]. To overcome the aforementioned limitation, various tools for the detection of periodontitis have been developed and introduced [2,3].
The use of saliva for oral-based diagnostics has proven to be easy to use for point of care (POC) application. Oral-based diagnostics have been developed to detect several pathologies, including oral cancer, human immunodeficiency virus infection, hepatitis C infection, and Ebola virus infection, with the advantages of being readily accessible and minimally invasive [4,5,6,7]. The ease of access and sampling of saliva containing inflammatory cytokines and microbial or viral infection provides potential possibilities for its use in the diagnosis of periodontitis [8].
It has been observed that there is an equilibrium in the interaction between the host and microorganisms when the periodontal apparatus is healthy [9]. This ecosystem can be affected by genetic factors of the host and environmental factors, including smoking [10]. The development of periodontal pathology is the outcome of several changes in the host or environmental state, indicating a possible relationship between the risk of periodontitis and susceptibility factors [11].
A previous study has demonstrated that microbial variables could show a discriminating potential in distinguishing subjects with periodontitis [3]. Dysbiosis of the oral microbiome can induce the immune reaction of the host, resulting in the release of various inflammatory mediators, including matrix metalloproteinase-8 (MMP-8), C-reactive protein (CRP), and secretory IgA (sIgA) [12,13,14].
MMP-8 (also known as type neutrophil collagenase or collagenase 2) has substrate specificity for type I collagen, accounting for periodontal extracellular matrix; thus, attention has been paid to MMP-8 in periodontitis [15,16]. Human MMP-8 is produced and secreted as a 55~80 kDa glycosylated inactive precursor form and is activated when cleaved by extracellular proteinases, resulting in the removal of the prodomain [17]. Inactive proMMP-8 can be activated by the “cystein switch” depending on several factors, including bacterial proteinase, temperature, pH value, calcium ion, and oxygen radicals [18]. Various studies have demonstrated that active MMP-8(aMMP-8) is related to periodontal pathogenesis [19,20,21]. Meanwhile, it is expected that the amount of aMMP-8 will also appear high in individuals with high expression of MMP-8. Therefore, it is necessary to measure all forms of MMP-8 (proMMP-8, aMMP-8, and total MMP-8) to examine the relationship between MMP-8 and the severity of periodontitis.
A variety of these microbial profiles and inflammatory mediators in saliva have been investigated as diagnostic tools for the detection of periodontitis. Although this diagnostic method has shown potential for identifying periodontitis, some limitations have been reported for this diagnostic tool, which uses a single biomarker [8]. There was a study that showed an improvement of sensitivity and specificity in detection using a model that combined related biomarkers [22]. As periodontitis is a multifactorial disease, a model combining microbial profiles and inflammatory mediators involved in chronic infection might improve its diagnostic accuracy.
Recently, for the purpose of reflecting the severity, extent, and complexity of periodontal breakdown, a new classification of periodontitis has been suggested [23]: Stage I, initial periodontitis; Stage II, moderate periodontitis; Stage III, severe periodontitis with potential for additional tooth loss; Stage IV, advanced periodontitis with extensive tooth loss and potential for loss of dentition. However, little evidence has been accumulated in support of using salivary biomarkers for such a classification system.
The aim of this study is to verify alterations in salivary biomarkers, including microbial profiles and inflammatory mediators, according to periodontal status and to investigate a combined model for periodontitis in accordance with the new classification.
2. Materials and Methods
2.1. Subjects
This clinical study was approved by the Institutional Review Board of Seoul National University Dental Hospital (CRI19004), approved on 4 April 2019. The subjects in this study were recruited from July 2019 to December 2019. Upon receiving written consent, 121 human subjects, aged 18 years or older, were evaluated at the Department of Periodontology in the Seoul National University Dental Hospital. All subjects involved in this study were required to have 20 or more teeth. In addition, the exclusion criteria applied was as follows: (a) use of any antibiotics or anti-inflammatory drugs within 3 months of registration; (b) use of an immunosuppressant (methotrexate, leflunomide, tacrolimus, cyclosporin, azathioprine) or adrenal cortical hormone (oral or injection) within 3 months of registration; (c) having less than 20 teeth; (d) having uncontrolled hypertension or diabetes; (e) subjects who have serious cardiovascular disease, respiratory system disease, kidney disease, liver disease, digestive system disease, blood system disease, or neuropsychiatric disease; (f) subjects with hyperthyroidism or hypothyroidism; (g) women who were pregnant or planning to become pregnant; (h) subjects with autoimmune diseases; (i) subjects with a history or presence of malignant tumors in the jawbone; (j) subjects who have had a history of or are currently using drugs or alcohol abuse within one year; (k) subjects with other inflammatory diseases in the oral cavity besides periodontitis, such as stomatitis (including ulcerative, blistering, erosive) or oral cancer; (l) subjects with other inflammatory diseases in the oral cavity besides periodontitis, such as ulcers, simple herpes and shingles, or fungal or bacterial infections; (m) subjects whose participation was judged by the researcher to be inappropriate because their involvement may cause ethical problems or seriously affect the research results.
2.2. Clinical and Radiographic Examination
A consent form was signed by and obtained from each subject following a sufficient explanation of the study. To identify if the subject was suitable for this study, demographic information such as gender and date of birth was obtained, and the systemic conditions of the participants were also examined.
On the first visit, clinical examinations were performed, and the following parameters were recorded. The gingival index (GI) [24] and plaque index (PI) [25] were examined on the buccal and lingual surfaces of the teeth. Additionally, the probing pocket depth (PPD), gingival recession (GR), and clinical attachment level (CAL) were measured at the six sites around the tooth. The amount of tooth loss (TL), sites of bleeding on probing (BOP), tooth mobility (TM), and furcation involvement (FI) were recorded per tooth. Alveolar bone loss at the mesial and distal site of the tooth was measured with periapical radiographs. The subjects were classified as healthy or Stages I, II, III, or IV depending on the severity, extent, and complexity of their periodontitis [23].
2.3. Preparation of Solution for Saliva Storage
Approximately 0.1 M phenylmethylsulphonyl fluoride (PMSF) stock solution, dissolved in isopropanol, was stored at room temperature and 0.5 M ethylenediaminetetraacetic acid (EDTA) stock solution, dissolved in distilled water (DW), was refrigerated. The two stock solutions were stored independently due to the instability of PMSF when mixed with 1X phosphate-buffered saline (PBS).
2.4. Whole Saliva with Draining Method
Unstimulated saliva was collected with the participant’s head tilted slightly forward in a sitting position by drooling into a funnel-shaped test tube. The sampling was performed for 15 min and was stopped when the amount collected reached 5 mL. Subsequently, the saliva sample was placed on ice and supplemented with 1X PBS 4930 μL, 20 μL EDTA solution, and 50 μL PMSF stock solution; then, vortexing was performed. The samples were stored in a deep freezer at a temperature of −80 °C immediately after collection for the preservation of biomarkers.
2.5. Saliva Collection for Oral Microbial Identification
After collecting GCF, subjects took another break for 5 min. The subjects gargled and rinsed with gargle solution (EasyperiO kit, YD Life Science Company, Gyeonggi-do, Korea) for 30 s and then spit it into the sample container. The sample container cap was closed tightly. Samples were stored in a refrigerator (4 °C) and transported to the analytical company (YD Life Science company, Gyeonggi-do, Korea) on dry ice.
2.6. Protein Biomarker Assays
Protein biomarker levels were detected with enzyme-linked immunosorbent assays (ELISAs) for measurement of active MMP-8 (Human MMP-8 activity assay, QuickZyme Biosciences B.V., Leiden, The Netherlands), total MMP-8 (Human MMP-8 ELISA Kit, RayBiotech, Inc., Norcross, GA, USA), CRP (Salivary C-Reactive Protein ELISA Kit, Salimetrics, LLC, State College, PA, USA), and sIgA (Secretory IgA ELISA Kit, Demeditec Diagnostics GmbH, Kiel, Germany), according to the manufacturers’ protocols. The total MMP-8 ELISA Kit is a sandwich method, and the active MMP-8 ELISA Kit is a method of indirectly measuring the amount of active MMP-8 by measuring the chromogenic substrate that is cleaved by the active detection enzyme activated by the active MMP-8. Pro-MMP-8 was calculated by subtracting active MMP-8 from total MMP-8. Pure recombinant human MMP-8 protein (aa 21–467; Sino Biological, Cat. No. 10254-HNAH, Beijing, China) was used as a positive control. Measurements were performed in duplicate to ensure repeatability of the experimental analysis.
2.7. Detection of Microbial Profiles
The samples were analyzed by quantitative analysis using multiplex-quantity real-time polymerase chain reaction (PCR) according to the manufacturer’s protocol (EasyPerio; YD Life Science, Gyeonggi-do, Korea) for 15 oral pathogenic bacteria: Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans, Aa), Porphyromonas gingivalis (P. gingivalis, Pg), Tannerella forsythia (T. forsythia, Tf), Treponema denticola (T. denticola, Td), Fusobacterium nucleatum (F. nucleatum, Fn), Prevotella intermedia (P. intermedia, Pi), Porphyromonas nigrescens (P. nigrescens, Pn), Parvimonas micra (P. micra, Pm), Campylobacter rectus (C. rectus, Cr), Eubacterium nodatum (E. nodatum, En), Eikenella corrodens (E. corrodens, Ec), Streptococcus mutans (S. mutans, Smu), Staphylococcus aureus (S. aureus, Sa), Enterococcus faecalis (E. Faecalis, Ef), and Actinomyces viscosus (A. viscosus, Av). Patient-based microbial data was analyzed. Sequences of the primers and probes used for real-time PCR are shown in Table 1. To obtain a standard curve for each target, plasmid DNA of each strain at a concentration of 104 copies/mL was prepared, and PCR was performed. The copy value for each strain was calculated by substituting the Ct value of each bacteria into a standard curve. Measurements were performed in duplicate to ensure repeatability of the experimental analysis.
2.8. Sample Size Calculation
The sample size was established according to the previous study by Mauramo M et al. [26], reporting the diagnostic performance of MMP-8 for periodontitis with an AUC value of 0.67. With a 95% confidence interval and 80% power, the sample size was calculated as a total of 108 subjects [27]. Give a 10% drop-out rate, 30 subjects were to be collected for each group of periodontitis.
2.9. Statistical Analysis
Continuous data were represented with means and standard deviation for each subject group. Group comparisons were made using one-way ANOVA in SPSS version 17 (IBM Software, Armonk, NY, USA). Dichotomized data were represented with a number and percentage for each subject group. Group comparisons were made with Fisher’s exact tests. Differences were considered statistically significant when p-values < 0.05. Seven diagnostic models of periodontitis (Model PD), namely, Stage I periodontitis (Model PD-I), Stage II periodontitis (Model PD-II), Stage III periodontitis (Model PD-III), Stage IV periodontitis (Model PD-IV), periodontitis above Stage I (Stages II, III, and IV; Model PA-I), and periodontitis above Stage II (Stages III and IV; Model PA-II), were constructed according to the concentration of proteins and microbial profiles based on a forward stepwise logistic regression analysis using SPSS statistics software (version 21.0, IBM Software, Armonk, NY, USA). The diagnostic models were evaluated by sensitivity, specificity, and ROC curve analysis using Excel (Microsoft 365, Redmond, WA, USA).
3. Results
3.1. Demographic and Clinical Characteristics of Subjects
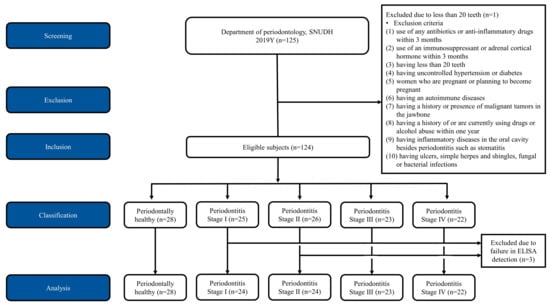
Thirty-eight male (30.4%) and eighty-seven female (69.6%) subjects, ranging in age from 20 to 79 years, were enrolled in the study. Following the recording of clinical and radiographic parameters, the subjects were allocated into the five groups of periodontal status. One subject was excluded due to the lack of teeth. The data of three subjects were excluded because the proteins in their whole-saliva samples, including active MMP-8, pro-MMP-8, and total MMP-8, did not show detectable levels. A total of 121 subjects were included in the final analysis (Figure 1). Analysis of the data obtained from the healthy (n = 28) and periodontitis populations (n = 93; Stage I: 24, Stage II: 24, Stage III: 23, and Stage IV: 22) was performed in this study.
Figure 1.
Consolidated standards of reporting trials (CONSORT) flow diagram of the study. 125 subjects were screened and one subject was excluded due to lack of teeth. Finally 124 subjects were included in this study and allocated into the five groups of periodontal status including healthy, periodontitis stage I, II, III, and IV. Three subjects were excluded in the analysis due to failure in enzyme-linked immunosorbent assay (ELISA) dectection. A total 121 subjects were included in the final analysis.
The demographic data (Table 2) for systemic disease, including osteoporosis and hepatitis, were balanced among the five groups. However, age, sex, hypertension, diabetes mellitus, and smoking were statistically and significantly different among the groups. The subjects with hypertension, diabetes, and smoking were not found in the periodontally healthy group; however, they were significantly associated with Stage III and Stage IV periodontitis. In addition, the older participants were significantly found to have higher stages of periodontitis.
Table 2.
Demographic characteristics, clinical periodontal parameters, salivary biomarkers, and planktonic bacteria of each group (mean ± standard deviation).
Dental and periodontal data (Table 2) were significantly different among the five groups for mean GR (0.11 to 0.65; p < 0.001), mean PPD (2.36 to 2.83 mm; p = 0.003), mean CAL (0.24 to 1.25 mm; p < 0.001), mean MBL (0.077 to 2.34 mm; p < 0.001), number of furcation-involved teeth (0 to 0.73; p < 0.001), number of immobile teeth (24.09 to 27.25; p < 0.001), number of mobile teeth with Grade 1 (0 to 1.64; p < 0.001), number of mobile teeth with Grade 2 (0 to 0.27; p = 0.001), and number of mobile teeth with Grade 3 (0 to 0.23; p = 0.022).
3.2. Inflammatory Mediators and Microbial Profiles in Saliva
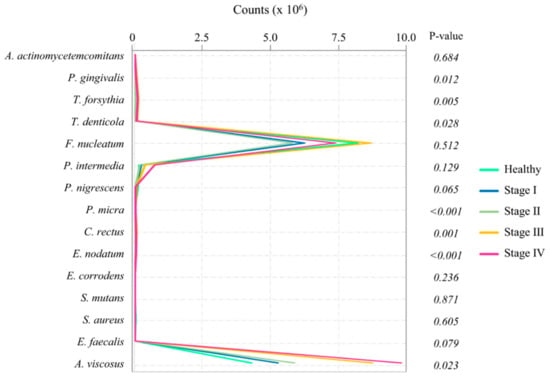
The data of proteins are shown in Table 2. The levels of protein concentrations of active MMP-8 (p < 0.001), pro-MMP-8 (p < 0.001), total MMP-8 (p < 0.001), and sIgA (p = 0.012) showed significant differences among the groups. The overall microbial profiles are presented in Figure 2. The level of P. gingivalis (p = 0.012), T. forsythia (p = 0.005), T. denticola (p = 0.028), P. micra (p < 0.001), C. rectus (p = 0.001), E. nodatum (p < 0.001), and A. viscosus (p = 0.023) showed significant differences among the groups.
3.3. Construction of Diagnostic Models Based on Inflammatory Mediators in Saliva
Diagnostic models of periodontitis (Model PD), Stage I periodontitis (Model PD-I), Stage II periodontitis (Model PD-II), Stage III periodontitis (Model PD-III), Stage IV periodontitis (Model PD-IV), periodontitis above Stage I (Stages II, III, and IV; Model PA-I), and periodontitis above Stage II (Stages III, and IV; Model PA-II) were constructed using the proteins and microbial profiles that showed significant differences.
The Model PD mathematical formula, with an accuracy of 0.802 (Table 3), was constructed to discriminate the periodontitis groups from the healthy group using logistic regression analysis (Equation (1)). This model showed high sensitivity (1.000), but low specificity (0.143) (Table 3).
The Model PD-I mathematical formula, with an accuracy of 0.785 (Table 3), was constructed to diagnose the Stage I periodontitis group from the healthy group and Stage II, III, and IV periodontitis patients using logistic regression analysis (Equation (2)). This model showed high specificity (0.969) but low sensitivity (0.042) (Table 3).
The Model PD-II mathematical formula, with an accuracy of 0.760 (Table 3), was constructed to diagnose the Stage II periodontitis group from the healthy group and Stage I, III, and IV periodontitis patients using logistic regression analysis (Equation (3)). This model showed high specificity (0.948) but low sensitivity (0.000) (Table 3).
The Model PD-III mathematical formula, with an accuracy of 0.835 (Table 3), was constructed to diagnose the Stage III periodontitis group from the healthy group and Stage I, II, and IV periodontitis patients using logistic regression analysis (Equation (4)). This model showed high specificity (0.959) but low sensitivity (0.304) (Table 3).
The Model PD-IV mathematical formula, with an accuracy of 0.860 (Table 3), was constructed to diagnose the Stage IV periodontitis group from the healthy group and Stage I, II, and III periodontitis patients using logistic regression analysis (Equation (5)). This model showed high specificity (0.980) but low sensitivity (0.318) (Table 3).
The Model PA-I mathematical formula, with an accuracy of 0.736 (Table 3), was constructed to discriminate the Stage II, III, and IV periodontitis groups from the healthy group and Stage I periodontitis patients using logistic regression analysis (Equation (6)). This model showed improved sensitivity (0.754) and specificity (0.712) (Table 3).
The Model PA-II mathematical formula, with an accuracy of 0.826 (Table 3), was constructed to discriminate the Stage III and IV periodontitis groups from the healthy group and Stage I and II periodontitis patients using logistic regression analysis (Equation (7)). This model showed good specificity (0.756) and sensitivity (0.868) (Table 3).
3.4. Validation of Multianalyte Models
Among the seven models, Model PA-I and Model PA-II, which showed good sensitivity and specificity, were further investigated with the ROC curves (Figure 3).
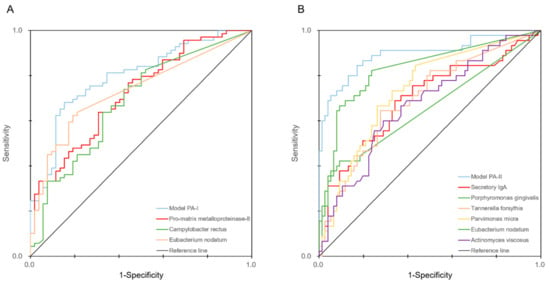
Figure 3.
ROC curves of the PA-I (A) and PA-II (B) models. The ROC curves of the biomarkers that were significantly related and included in each model are also included in each figure.
It was found that the models combining salivary biomarkers and microbial profiles were useful for discriminating periodontal status. The AUC values of pro-MMP-8, C. rectus, and E. nodatum were 0.720, 0.685, and 0.733, respectively. In contrast, the AUC value of Model PA-I, which combined the markers, was 0.796. The AUC values of sIgA, P. gingivalis, T. forsythia, P. micra, E. nodatum, and A. viscosus were 0.695, 0.653, 0.694, 0.736, 0.831, and 0.673, respectively, while Model PA-II, which combined them, showed an AUC value of 0.894.
4. Discussion
In this study, we built the seven diagnostic models for periodontitis by combining meaningful salivary biomarkers (including inflammatory mediators and microbial profiles) that significantly changed with the stages of periodontitis. Among the seven models, PA-I and PA-II were both highly sensitive and specific compared with PD-I, -II, -III, and -IV models. In Model PA-I, E. nodatum and C. rectus were included; these bacteria belong to the orange complex, which has a role in linking early colonizing and pathogenic bacteria of periodontitis. Therefore, it is thought that these bridging species are very important in the diagnosis of periodontitis with Stages II, III, and IV of the disease. Meanwhile, in Model PA-II, the purple complex (such as A. viscosus), orange complex (such as P. micra and E. nodatum), and red complex (such as P. gingivalis and T. forsythia) were included. The high accuracy of Model PA-II means that overall planktonic bacterial species can increase when the severity of periodontitis increases because various species consisting of early colonizer, bridging species, and pathogenic bacteria are included. Simple counting of bacteria also showed that there were more groups of streptococcus and A. viscosus in Stage IV, meaning the more severe the stage of periodontitis is, the more overall bacteria there are.
To our knowledge, there is little literature to validate a new classification system of periodontitis in the development of a diagnostic method, including salivary biomarkers. In 2017, the new classification system for case definitions of periodontitis was developed and suggested based on cumulative studies for almost 20 years [28]. In the workshop, three obviously different forms of periodontitis based on pathophysiology were categorized: necrotizing periodontitis, periodontitis as a direct manifestation of systemic disease, and periodontitis. Herein, we included the latter form. The periodontitis stage increased according to the severity, complexity, extent, and distribution of the disease. Our prediction models for periodontitis showed high accuracy based on this new classification system by using limited amounts of significant biomarkers.
In this study, we included both pro- and active forms of MMP-8. Several studies found that the active forms of MMP could distinguish periodontitis [19,29,30,31,32,33,34]; however, in this study, two forms were significantly different among all stages of periodontitis. The increase was considered to reflect the increased leakage of all types of MMP-8 according to the severity of periodontitis. Interestingly, CRP did not show a significant difference, which was not consistent with other studies [35,36]. This might be because the detective capacity of ELISA is not sensitive enough to detect the changes in the level of CRP [37,38]. Therefore, to monitor the level of CRP, more sensitive techniques should be used.
Contrary to our expectations, all periodontopathogenic bacteria were not detected in the saliva in periodontitis patients. This could be a result of the fact that almost all the patients showed periodontitis in a “localized” form, and the amount of planktonic bacteria was too low to be detected. Further studies should be performed in patients with “generalized” periodontitis.
SIgA indicates adaptive immunity and is widely used for a diagnosis of periodontitis. Mesa et al. reported similar results showing sIgA was not significantly different between healthy patients and periodontitis groups [39]. Chronic inflammation increased cortisol levels and, inversely, decreased sIgA [40]. In the present study, sIgA seemed to show higher levels in Stages III and IV of periodontitis compared with other groups, but there were no statistical differences.
In spite of the limitations of the cross-sectional study, our findings suggest that whole saliva might be used as a diagnostic tool for periodontitis. Additionally, the proper selection of biomarkers in whole saliva is important in order to increase the sensitivity and specificity of the diagnosis of periodontitis [41,42]. On the other hand, this method had limitations in the early diagnosis of Stage I. Other studies also showed similar limitations, where the screening test could not distinguish early-stage periodontitis [43]. By definition, Stage I periodontitis is a bridge between gingivitis and periodontitis; thus, it can show lower levels of biomarkers than the severe forms. Other researchers have tried to discriminate gingivitis and periodontitis through macrophage inflammatory protein-1α [44,45]. To overcome the problem, it is necessary to find additional biomarkers or develop more sensitive techniques for early detection.
The diagnosis of periodontitis with clinical parameters is a very effective tool, but it is time-consuming and labor-intensive. The evaluation of clinical parameters is somewhat difficult to standardize and cannot monitor the real-time changes in periodontal disease progression [41,42]. Therefore, the method of whole-saliva analysis could be an easier and simpler diagnostic tool for the detection of periodontitis. Additionally, salivary biomarkers can be a very prospective screening tool when one considers the high correlations between periodontitis and systemic disease [29,46,47].
5. Conclusions
This study can contribute to screening for periodontitis based on salivary biomarkers. Two potential diagnosis models, PA-I for the diagnosis of Stage II, III, and IV periodontitis and PA-II for the diagnosis of Stage III and IV periodontitis, showed the highest performance with biomarkers in whole saliva.
Author Contributions
Conceptualization: J.L. and K.-T.K.; formal analysis: J.L., J.-B.L. and J.-S.A.; investigation: J.L., K.-T.K., J.S.K. and E.K.; data curation: H.-Y.S., M.J.S. and L.L.; methodology: I.-C.R. and Y.-M.L.; visualization: J.-B.L. and J.-S.A.; project administration: J.L. and K.-T.K.; funding acquisition: K.-T.K.; writing—original draft preparation: J.L. and J.-B.L.; writing—review and editing: H.-Y.S., M.J.S., L.L., I.-C.R., Y.-M.L., K.-T.K., J.-S.A., J.S.K. and E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Biotechnology Development Project—Advanced New Biomaterials (20000580, “Development of a Point of Care Testing System for High Sensitivity Detection of Biomarkers in Saliva”), funded by the Ministry of Trade, Industry, and Energy (MOTIE, Korea).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yao, Y.; Jiao, K.; Zhang, J.; Zheng, X.; Wu, F.; Hu, X.; Li, J.; Yu, Z.; Zhang, G.; et al. Relationship between Gingival Crevicular Fluid Microbiota and Cytokine Profile in Periodontal Host Homeostasis. Front. Microbiol. 2017, 8, 2144. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, T.G.; Heller, D.; Silva-Boghossian, C.M.; Cotton, S.L.; Paster, B.J.; Colombo, A.P. Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 2014, 41, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Holguin, A.; Gutierrez, M.; Portocarrero, N.; Rivas, P.; Baquero, M. Performance of OraQuick Advance Rapid HIV-1/2 Antibody Test for detection of antibodies in oral fluid and serum/plasma in HIV-1+ subjects carrying different HIV-1 subtypes and recombinant variants. J. Clin. Virol. 2009, 45, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Louis, J.F.; Huang, J.Y.; Nebie, Y.K.; Koivogui, L.; Jayaraman, G.; Abiola, N.; Vansteelandt, A.; Worrel, M.C.; Shang, J.; Murphy, L.B.; et al. Implementation of broad screening with Ebola rapid diagnostic tests in Forecariah, Guinea. Afr. J. Lab. Med. 2017, 6, 484. [Google Scholar] [CrossRef]
- Lee, S.R.; Kardos, K.W.; Schiff, E.; Berne, C.A.; Mounzer, K.; Banks, A.T.; Tatum, H.A.; Friel, T.J.; Demicco, M.P.; Lee, W.M.; et al. Evaluation of a new, rapid test for detecting HCV infection, suitable for use with blood or oral fluid. J. Virol. Methods 2011, 172, 27–31. [Google Scholar] [CrossRef]
- Zimmermann, B.G.; Wong, D.T. Salivary mRNA targets for cancer diagnostics. Oral Oncol. 2008, 44, 425–429. [Google Scholar] [CrossRef]
- Leppilahti, J.M.; Harjunmaa, U.; Järnstedt, J.; Mangani, C.; Hernández, M.; Tervahartiala, T.; Lopez, R.; Ashorn, U.; Ashorn, P.; Gieselmann, D.R.; et al. Diagnosis of Newly Delivered Mothers for Periodontitis with a Novel Oral-Rinse aMMP-8 Point-of-Care Test in a Rural Malawian Population. Diagnostics 2018, 8, 67. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef]
- Ulloa, P.C.; van der Veen, M.H.; Krom, B.P. Review: Modulation of the oral microbiome by the host to promote ecological balance. Odontology 2019, 107, 437–448. [Google Scholar] [CrossRef]
- Kinane, D.F.; Attström, R. Advances in the pathogenesis of periodontitis. Group B consensus report of the fifth European Workshop in Periodontology. J. Clin. Periodontol. 2005, 32 (Suppl. 6), 130–131. [Google Scholar] [CrossRef] [PubMed]
- De Morais, E.F.; Pinheiro, J.C.; Leite, R.B.; Santos, P.P.A.; Barboza, C.A.G.; Freitas, R.A. Matrix metalloproteinase-8 levels in periodontal disease patients: A systematic review. J. Periodontal Res. 2018, 53, 156–163. [Google Scholar] [CrossRef]
- Rangbulla, V.; Nirola, A.; Gupta, M.; Batra, P. Salivary IgA, Interleukin-1β and MMP-8 as Salivary Biomarkers in Chronic Periodontitis Patients. Chin. J. Dent. Res. 2017, 20, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Shojaee, M.; Golpasha, M.F.; Maliji, G.; Bijani, A.; Mir, S.M.A.; Kani, S.N.M. C-reactive protein levels in patients with periodontal disease and normal subjects. Int. J. Mol. Cell. Med. 2013, 2, 151–155. [Google Scholar]
- Rathnayake, N.; Gustafsson, A.; Norhammar, A.; Kjellstrom, B.; Klinge, B.; Ryden, L.; Tervahartiala, T.; Sorsa, T.; Group, P.S. Salivary Matrix Metalloproteinase-8 and -9 and Myeloperoxidase in Relation to Coronary Heart and Periodontal Diseases: A Subgroup Report from the PAROKRANK Study (Periodontitis and Its Relation to Coronary Artery Disease). PLoS ONE 2015, 10, e0126370. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Gupta, N.D.; Gupta, A.; Khan, S.; Bansal, N. Role of salivary matrix metalloproteinase-8 (MMP-8) in chronic periodontitis diagnosis. Front. Med. 2015, 9, 72–76. [Google Scholar] [CrossRef]
- Hanemaaijer, R.; Sorsa, T.; Konttinen, Y.T.; Ding, Y.; Sutinen, M.; Visser, H.; van Hinsbergh, V.W.; Helaakoski, T.; Kainulainen, T.; Ronka, H.; et al. Matrix metalloproteinase-8 is expressed in rheumatoid synovial fibroblasts and endothelial cells. Regulation by tumor necrosis factor-alpha and doxycycline. J. Biol. Chem. 1997, 272, 31504–31509. [Google Scholar] [CrossRef]
- Franco, C.; Patricia, H.R.; Timo, S.; Claudia, B.; Marcela, H. Matrix Metalloproteinases as Regulators of Periodontal Inflammation. Int. J. Mol. Sci. 2017, 18, 440. [Google Scholar] [CrossRef]
- Alassiri, S.; Parnanen, P.; Rathnayake, N.; Johannsen, G.; Heikkinen, A.M.; Lazzara, R.; van der Schoor, P.; van der Schoor, J.G.; Tervahartiala, T.; Gieselmann, D.; et al. The Ability of Quantitative, Specific, and Sensitive Point-of-Care/Chair-Side Oral Fluid Immunotests for aMMP-8 to Detect Periodontal and Peri-Implant Diseases. Dis. Markers 2018, 2018, 1306396. [Google Scholar] [CrossRef]
- Schmalz, G.; Kummer, M.K.; Kottmann, T.; Rinke, S.; Haak, R.; Krause, F.; Schmidt, J.; Ziebolz, D. Association of chairside salivary aMMP-8 findings with periodontal risk assessment parameters in patients receiving supportive periodontal therapy. J. Periodontal Implant Sci. 2018, 48, 251–260. [Google Scholar] [CrossRef]
- Sorsa, T.; Tjaderhane, L.; Konttinen, Y.T.; Lauhio, A.; Salo, T.; Lee, H.M.; Golub, L.M.; Brown, D.L.; Mantyla, P. Matrix metalloproteinases: Contribution to pathogenesis, diagnosis and treatment of periodontal inflammation. Ann. Med. 2006, 38, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, H.; Qiao, Y.; Yang, J.; Shu, J.; Zhang, J.; Zhang, Z.; He, J.; Li, Z. Salivary Glycopatterns as Potential Biomarkers for Screening of Early-Stage Breast Cancer. EBioMedicine 2018, 28, 70–79. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S149–S161. [Google Scholar] [CrossRef] [PubMed]
- Loe, H.; Silness, J. Periodontal disease in pregnancy. I. prevalence and severity. Acta Odontol. Scand. 1963, 21, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Silness, J.; Loe, H. Periodontal disease in pregnancy. II. correlation between oral hygiene and periodontal condtion. Acta Odontol. Scand. 1964, 22, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Mauramo, M.; Ramseier, A.M.; Mauramo, E.; Buser, A.; Tervahartiala, T.; Sorsa, T.; Waltimo, T. Associations of oral fluid MMP-8 with periodontitis in Swiss adult subjects. Oral Dis. 2018, 24, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.E.; Bachmann, L.M.; Jaeschke, R. A readers’ guide to the interpretation of diagnostic test properties: Clinical example of sepsis. Intensive Care Med. 2003, 29, 1043–1051. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. 1), S159–S172. [Google Scholar] [CrossRef]
- Gursoy, U.K.; Könönen, E.; Pradhan-Palikhe, P.; Tervahartiala, T.; Pussinen, P.J.; Suominen-Taipale, L.; Sorsa, T. Salivary MMP-8, TIMP-1, and ICTP as markers of advanced periodontitis. J. Clin. Periodontol. 2010, 37, 487–493. [Google Scholar] [CrossRef]
- Sorsa, T.; Alassiri, S.; Grigoriadis, A.; Räisänen, I.T.; Pärnänen, P.; Nwhator, S.O.; Gieselmann, D.R.; Sakellari, D. Active MMP-8 (aMMP-8) as a Grading and Staging Biomarker in the Periodontitis Classification. Diagnostics 2020, 10, 61. [Google Scholar] [CrossRef]
- Yucel, Z.P.K.; Afacan, B.; Emingil, G.; Tervahartiala, T.; Kose, T.; Sorsa, T. Local and systemic levels of aMMP-8 in gingivitis and stage 3 grade C periodontitis. J. Periodontal Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sorsa, T.; Bacigalupo, J.; Könönen, M.; Pärnänen, P.; Räisänen, I.T. Host-Modulation Therapy and Chair-Side Diagnostics in the Treatment of Peri-Implantitis. Biosensors 2020, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Lähteenmäki, H.; Umeizudike, K.A.; Heikkinen, A.M.; Räisänen, I.T.; Rathnayake, N.; Johannsen, G.; Tervahartiala, T.; Nwhator, S.O.; Sorsa, T. aMMP-8 Point-of-Care/Chairside Oral Fluid Technology as a Rapid, Non-Invasive Tool for Periodontitis and Peri-Implantitis Screening in a Medical Care Setting. Diagnostics 2020, 10, 562. [Google Scholar] [CrossRef] [PubMed]
- Al-Majid, A.; Alassiri, S.; Rathnayake, N.; Tervahartiala, T.; Gieselmann, D.R.; Sorsa, T. Matrix Metalloproteinase-8 as an Inflammatory and Prevention Biomarker in Periodontal and Peri-Implant Diseases. Int. J. Dent. 2018, 2018, 7891323. [Google Scholar] [CrossRef]
- Podzimek, S.; Vondrackova, L.; Duskova, J.; Janatova, T.; Broukal, Z. Salivary Markers for Periodontal and General Diseases. Dis. Markers 2016, 2016, 9179632. [Google Scholar] [CrossRef]
- Shimada, Y.; Komatsu, Y.; Ikezawa-Suzuki, I.; Tai, H.; Sugita, N.; Yoshie, H. The effect of periodontal treatment on serum leptin, interleukin-6, and C-reactive protein. J. Periodontol. 2010, 81, 1118–1123. [Google Scholar] [CrossRef]
- Christodoulides, N.; Floriano, P.N.; Miller, C.S.; Ebersole, J.L.; Mohanty, S.; Dharshan, P.; Griffin, M.; Lennart, A.; Ballard, K.L.; King, C.P., Jr.; et al. Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Ann. N. Y. Acad. Sci. 2007, 1098, 411–428. [Google Scholar] [CrossRef]
- Wu, Y.C.; Ning, L.; Tu, Y.K.; Huang, C.P.; Huang, N.T.; Chen, Y.F.; Chang, P.C. Salivary biomarker combination prediction model for the diagnosis of periodontitis in a Taiwanese population. J. Formos. Med. Assoc. 2018, 117, 841–848. [Google Scholar] [CrossRef]
- Mesa, F.; Magán-Fernández, A.; Muñoz, R.; Papay-Ramírez, L.; Poyatos, R.; Sánchez-Fernández, E.; Galindo-Moreno, P.; Rodríguez-Barranco, M. Catecholamine metabolites in urine, as chronic stress biomarkers, are associated with higher risk of chronic periodontitis in adults. J. Periodontol. 2014, 85, 1755–1762. [Google Scholar] [CrossRef]
- Hucklebridge, F.; Clow, A.; Evans, P. The relationship between salivary secretory immunoglobulin A and cortisol: Neuroendocrine response to awakening and the diurnal cycle. Int. J. Psychophysiol. 1998, 31, 69–76. [Google Scholar] [CrossRef]
- Giannobile, W.V. Salivary diagnostics for periodontal diseases. J. Am. Dent. Assoc. 2012, 143, 6s–11s. [Google Scholar] [CrossRef] [PubMed]
- Korte, D.L.; Kinney, J. Personalized medicine: An update of salivary biomarkers for periodontal diseases. Periodontology 2000 2016, 70, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, M.J.L.; Teeuw, W.J.; Bizzarro, S.; Muris, J.; Su, N.; Nicu, E.A.; Nazmi, K.; Bikker, F.J.; Loos, B.G. A rapid, non-invasive tool for periodontitis screening in a medical care setting. BMC Oral Health 2019, 19, 87. [Google Scholar] [CrossRef]
- Kim, J.J.; Kim, C.J.; Camargo, P.M. Salivary biomarkers in the diagnosis of periodontal diseases. J. Calif. Dent. Assoc. 2013, 41, 119–124. [Google Scholar] [PubMed]
- Syndergaard, B.; Al-Sabbagh, M.; Kryscio, R.J.; Xi, J.; Ding, X.; Ebersole, J.L.; Miller, C.S. Salivary biomarkers associated with gingivitis and response to therapy. J. Periodontol. 2014, 85, e295–e303. [Google Scholar] [CrossRef] [PubMed]
- Lahdentausta, L.S.J.; Paju, S.; Mäntylä, P.; Buhlin, K.; Tervahartiala, T.; Pietiäinen, M.; Alfthan, H.; Nieminen, M.S.; Sinisalo, J.; Sorsa, T.; et al. Saliva and serum biomarkers in periodontitis and coronary artery disease. J. Clin. Periodontol. 2018, 45, 1045–1055. [Google Scholar] [CrossRef]
- Mirrielees, J.; Crofford, L.J.; Lin, Y.; Kryscio, R.J.; Dawson, D.R., 3rd; Ebersole, J.L.; Miller, C.S. Rheumatoid arthritis and salivary biomarkers of periodontal disease. J. Clin. Periodontol. 2010, 37, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).