Predicting Clinical Outcome in Acute Ischemic Stroke Using Parallel Multi-Parametric Feature Embedded Siamese Network
Abstract
:1. Introduction
2. Literature Review
2.1. Dichotomized Output
2.2. Non-Dichotomized Output
3. Methodology
3.1. Data
3.2. Basic Framework
3.2.1. Preprocessing
3.2.2. Data Augmentation
3.2.3. Pair Creation for Training
Pairs of Samples from Same Classes
Pairs of Samples from Dissimilar Classes
3.2.4. Classification Model Architecture
Parallel Multi-Parametric Feature Embedding
Distance Metric and Loss Function
| Algorithm 1. Algorithm of PMFE-SN in leave one cross-out fold |
| is the number of samples in the training set . |
| M is the number of classes in the training set. |
| is the number of samples belonging to class . |
| , the set of class labels. |
| Training Phase |
| Input: Training set , where each ∈ L |
| Method: |
| 1: Identify I, representing the largest number of samples amongst all classes. |
| 2: Augment each class m with I−nm number of new samples in the training set. |
| 3: Create unique pairs of samples from class m where and put in set . |
| 4: Assign label 1 to each pair in set SP. |
| 5: Create unique number of pairs of dissimilar class labels from set L and put in set LP. |
| 6: Create unique pairs of samples for each dissimilar class label pairs in LP and put in set DP. |
| 7: Assign label 0 to each pair in set DP. |
| 8: Let such that , where pi is the ith pair of training and bi ∈ {0,1} |
| 9: Split Train in ratio 7:3 for training and validation of PMFE-SN. |
| 10: Train PMFE-SN. |
| Testing Phase |
| Input: Initial Training set , where each ∈ L |
| Test Sample: |
| Method: |
| 1: Make pairs of testing sample with every original sample in X. |
| 2: Pass all pairs from trained PMFE-SN to get similarity of each pair. |
| 3: Class of sample in X with highest similarity with is the predicted class of . |
| 4: If there is more than 1 sample in X with highest similarity with , choose randomly amongst such samples and assign the class of randomly chosen sample to . |
3.3. Evaluation Metrics
4. Experimental Setup
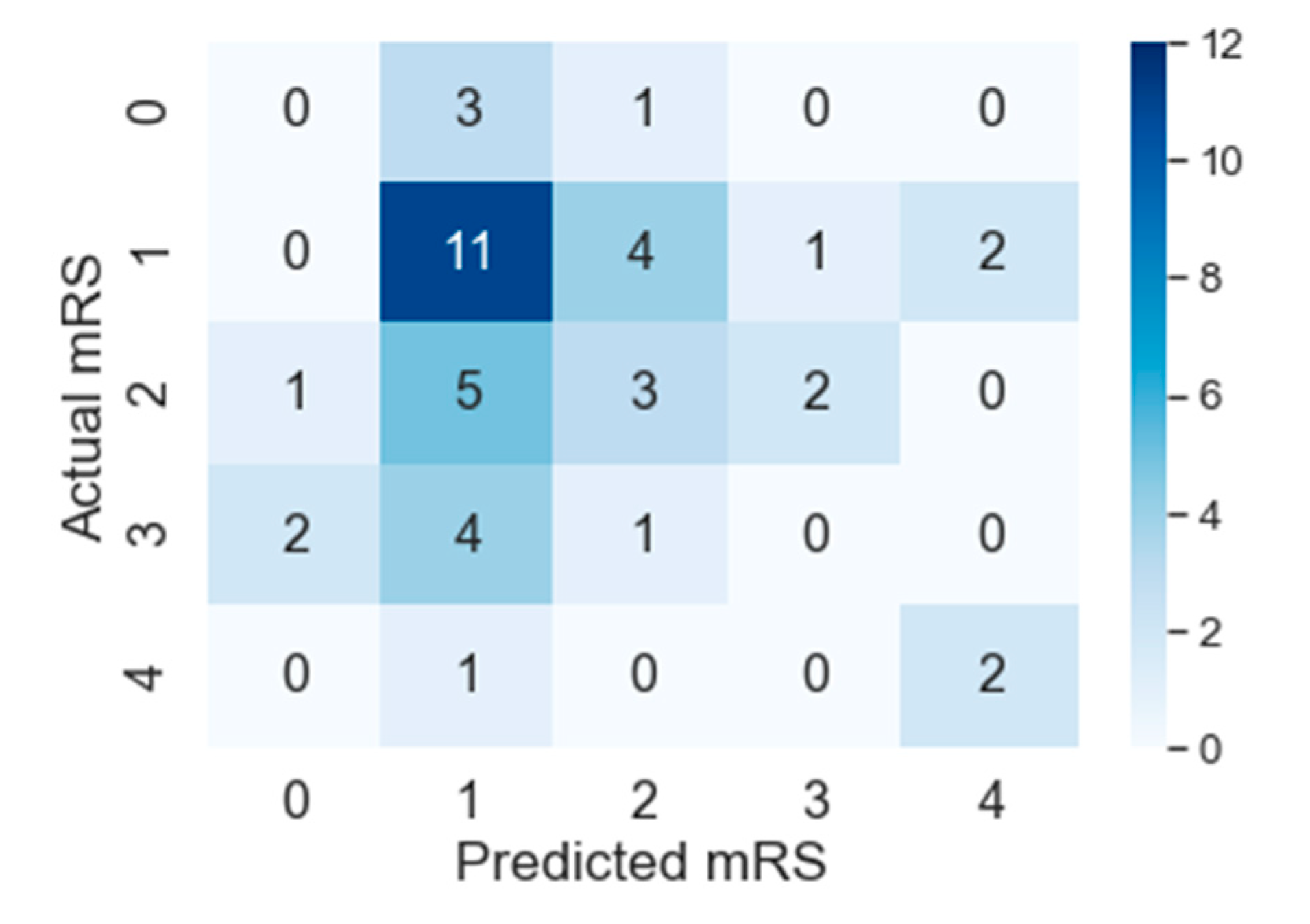
5. Results and Discussion
Effect of Augmentation
6. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Feigin, V.L.; Norrving, B.; Mensah, G.A. Global Burden of Stroke. Circ. Res. 2017, 120, 439–448. [Google Scholar] [CrossRef] [PubMed]
- WHO EMRO. WHO Emro. Available online: http://www.emro.who.int/index.html (accessed on 27 December 2019).
- González, R.G.; Hirsch, J.A.; Lev, M.H.; Schaefer, P.W.; Schwamm, L.H. Acute Ischemic Stroke: Imaging and Intervention; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 9783540252641. [Google Scholar]
- El Tawil, S.; Muir, K.W. Thrombolysis and thrombectomy for acute ischaemic stroke. Clin. Med. J. R. Coll. Physicians Lond. 2017, 17, 161–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higashida, R.T.; Furlan, A.J.; Roberts, H.; Tomsick, T.; Connors, B.; Barr, J.; Dillon, W.; Warach, S.; Broderick, J.; Tilley, B.; et al. Trial design and reporting standards for intraarterial cerebral thrombolysis for acute ischemic stroke. J. Vasc. Interv. Radiol. 2003, 14, 945–946. [Google Scholar] [CrossRef]
- Ebinger, M.; Kufner, A.; Galinovic, I.; Brunecker, P.; Malzahn, U.; Nolte, C.H.; Endres, M.; Fiebach, J.B. Fluid-attenuated inversion recovery images and stroke outcome after thrombolysis. Stroke 2012, 43, 539–542. [Google Scholar] [CrossRef]
- Hung, C.Y.; Chen, W.C.; Lai, P.T.; Lin, C.H.; Lee, C.C. Comparing deep neural network and other machine learning algorithms for stroke prediction in a large-scale population-based electronic medical claims database. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Jeju Island, Korea, 11–15 July 2017; pp. 3110–3113. [Google Scholar]
- Feng, R.; Badgeley, M.; Mocco, J.; Oermann, E.K. Deep learning guided stroke management: A review of clinical applications. J. Neurointerv. Surg. 2018, 10, 358–361. [Google Scholar] [CrossRef]
- Kamal, H.; Lopez, V.; Sheth, S.A. Machine learning in acute ischemic stroke neuroimaging. Front. Neurol. 2018, 9, 945. [Google Scholar] [CrossRef]
- Rekik, I.; Allassonnière, S.; Carpenter, T.K.; Wardlaw, J.M. Medical image analysis methods in MR/CT-imaged acute-subacute ischemic stroke lesion: Segmentation, prediction and insights into dynamic evolution simulation models. A critical appraisal. NeuroImage Clin. 2012, 1, 164–178. [Google Scholar] [CrossRef] [Green Version]
- Maier, O.; Menze, B.H.; von der Gablentz, J.; Häni, L.; Heinrich, M.P.; Liebrand, M.; Winzeck, S.; Basit, A.; Bentley, P.; Chen, L.; et al. ISLES 2015-A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal. 2017, 35, 250–269. [Google Scholar] [CrossRef] [Green Version]
- Winzeck, S.; Hakim, A.; McKinley, R.; Pinto, J.A.A.D.S.R.; Alves, V.; Silva, C.; Pisov, M.; Krivov, E.; Belyaev, M.; Monteiro, M.; et al. ISLES 2016 and 2017-benchmarking ischemic stroke lesion outcome prediction based on multispectral MRI. Front. Neurol. 2018, 9, 679. [Google Scholar] [CrossRef]
- Ho, K.C.; Speier, W.; El-Saden, S.; Liebeskind, D.S.; Saver, J.L.; Bui, A.A.T.; Arnold, C.W. Predicting discharge mortality after acute ischemic stroke using balanced data. AMIA Annu. Symp. Proc. 2014, 2014, 1787–1796. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Bentley, P.; Ganesalingam, J.; Carlton Jones, A.L.; Mahady, K.; Epton, S.; Rinne, P.; Sharma, P.; Halse, O.; Mehta, A.; Rueckert, D. Prediction of stroke thrombolysis outcome using CT brain machine learning. NeuroImage Clin. 2014, 4, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fransen, P.S.S.; Beumer, D.; Berkhemer, O.A.; van den Berg, L.A.; Lingsma, H.; van der Lugt, A.; van Zwam, W.H.; van Oostenbrugge, R.J.; Roos, Y.B.W.E.M.; Majoie, C.B.; et al. MR CLEAN, a multicenter randomized clinical trial of endovascular treatment for acute ischemic stroke in the Netherlands: Study protocol for a randomized controlled trial. Trials 2014, 15, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Os, H.J.A.; Ramos, L.A.; Hilbert, A.; Van Leeuwen, M.; Van Walderveen, M.A.A.; Kruyt, N.D.; Dippel, D.W.J.; Steyerberg, E.W.; Van Der Schaaf, I.C.; Lingsma, H.F.; et al. Predicting outcome of endovascular treatment for acute ischemic stroke: Potential value of machine learning algorithms. Front. Neurol. 2018, 9, 784. [Google Scholar] [CrossRef]
- Xie, Y.; Jiang, B.; Gong, E.; Li, Y.; Zhu, G.; Michel, P.; Wintermark, M.; Zaharchuk, G. Use of gradient boosting machine learning to predict patient outcome in acute ischemic stroke on the basis of imaging, demographic, and clinical information. Am. J. Roentgenol. 2019, 212, 44–51. [Google Scholar] [CrossRef]
- Tang, T.Y.; Jiao, Y.; Cui, Y.; Zhao, D.L.; Zhang, Y.; Wang, Z.; Meng, X.P.; Yin, X.D.; Yang, Y.J.; Teng, G.J.; et al. Penumbra-based radiomics signature as prognostic biomarkers for thrombolysis of acute ischemic stroke patients: A multicenter cohort study. J. Neurol. 2020. [Google Scholar] [CrossRef]
- Lin, C.H.; Hsu, K.C.; Johnson, K.R.; Fann, Y.C.; Tsai, C.H.; Sun, Y.; Lien, L.M.; Chang, W.L.; Chen, P.L.; Lin, C.L.; et al. Evaluation of machine learning methods to stroke outcome prediction using a nationwide disease registry. Comput. Methods Programs Biomed. 2020, 190. [Google Scholar] [CrossRef]
- Wang, F.; Huang, Y.; Xia, Y.; Zhang, W.; Fang, K.; Zhou, X.; Yu, X.; Cheng, X.; Li, G.; Wang, X.; et al. Personalized risk prediction of symptomatic intracerebral hemorrhage after stroke thrombolysis using a machine-learning model. Ther. Adv. Neurol. Disord. 2020, 13. [Google Scholar] [CrossRef] [Green Version]
- Bacchi, S.; Zerner, T.; Oakden-Rayner, L.; Kleinig, T.; Patel, S.; Jannes, J. Deep Learning in the Prediction of Ischaemic Stroke Thrombolysis Functional Outcomes: A Pilot Study. Acad. Radiol. 2020, 27, e19–e23. [Google Scholar] [CrossRef]
- Wang, Q.; Reps, J.M.; Kostka, K.F.; Ryan, P.B.; Zou, Y.; Voss, E.A.; Rijnbeek, P.R.; Chen, R.; Rao, G.A.; Stewart, H.M.; et al. Development and validation of a prognostic model predicting symptomatic hemorrhagic transformation in acute ischemic stroke at scale in the OHDSI network. PLoS ONE 2020, 15, e226718. [Google Scholar] [CrossRef] [Green Version]
- Heo, J.N.; Yoon, J.G.; Park, H.; Kim, Y.D.; Nam, H.S.; Heo, J.H. Machine Learning-Based Model for Prediction of Outcomes in Acute Stroke. Stroke 2019, 50, 1263–1265. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Oishi, N.; Ishii, A.; Chihara, H.; Ogura, T.; Ishibashi, R.; Ando, M.; Sadamasa, N.; Ohta, T.; Kai, Y.; et al. Abstract TP83: Predicting Clinical Outcomes of Acute Ischemic Stroke Due to Large Vessel Occlusion: The Approach to Utilize High-dimensional Neuroimaging Data with Deep Learning. Stroke 2019, 50. [Google Scholar] [CrossRef]
- Hung, L.-C.; Sung, S.-F.; Hu, Y.-H. A Machine Learning Approach to Predicting Readmission or Mortality in Patients Hospitalized for Stroke or Transient Ischemic Attack. Appl. Sci. 2020, 10, 6337. [Google Scholar] [CrossRef]
- Mah, Y.H.; Nachev, P.; MacKinnon, A.D. Quantifying the Impact of Chronic Ischemic Injury on Clinical Outcomes in Acute Stroke with Machine Learning. Front. Neurol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kiik, M.; Peek, N.; Curcin, V.; Marshall, I.J.; Rudd, A.G.; Wang, Y.; Douiri, A.; Wolfe, C.D.; Bray, B. A systematic review of machine learning models for predicting outcomes of stroke with structured data. PLoS ONE 2020, 15, e0234722. [Google Scholar] [CrossRef]
- Asadi, H.; Dowling, R.; Yan, B.; Mitchell, P. Machine learning for outcome prediction of acute ischemic stroke post intra-arterial therapy. PLoS ONE 2014, 9, e88225. [Google Scholar] [CrossRef] [Green Version]
- Forkert, N.D.; Verleger, T.; Cheng, B.; Thomalla, G.; Hilgetag, C.C.; Fiehler, J. Multiclass support vector machine-based lesion mapping predicts functional outcome in ischemic stroke patients. PLoS ONE 2015, 10, e129569. [Google Scholar] [CrossRef]
- Mahmood, Q.; Basit, A. Prediction of ischemic stroke lesion and clinical outcome in multi-modal MRI images using random forests. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Berlin/Heidelberg, Germany, 2016; Volume 10154 LNCS, pp. 244–255. [Google Scholar]
- Egger, K.; Maier, O.; Reyes, M.; Wiest, R. ISLES: Ischemic Stroke Lesion Segmentation Challenge. 2016. Available online: http://www.isles-challenge.org/ISLES2017/ (accessed on 27 December 2019).
- Choi, Y.; Kwon, Y.; Lee, H.; Kim, B.J.; Paik, M.C.; Won, J.H. Ensemble of deep convolutional neural networks for prognosis of ischemic stroke. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Berlin/Heidelberg, Germany, 2016; Volume 10154 LNCS, pp. 231–243. [Google Scholar]
- Maier, O.; Handels, H. Predicting stroke lesion and clinical outcome with random forests. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Berlin/Heidelberg, Germany, 2016; Volume 10154 LNCS, pp. 219–230. [Google Scholar]
- Maier, O.; Wilms, M.; von der Gablentz, J.; Krämer, U.M.; Münte, T.F.; Handels, H. Extra Tree forests for sub-acute ischemic stroke lesion segmentation in MR sequences. J. Neurosci. Methods 2015, 240, 89–100. [Google Scholar] [CrossRef]
- Kabir, A.; Ruiz, C.; Alvarez, S.A.; Moonis, M. Predicting outcome of ischemic stroke patients using bootstrap aggregating with M5 model trees. In Proceedings of the HEALTHINF 2017-10th International Conference on Health Informatics; Part of 10th International Joint Conference on Biomedical Engineering Systems and Technologies, BIOSTEC 2017, Porto, Portugal, 21–23 February 2017; Volume 5, pp. 178–187. [Google Scholar]
- Samak, Z.A.; Clatworthy, P.; Mirmehdi, M. Prediction of Thrombectomy Functional Outcomes Using Multimodal Data. In Proceedings of the Communications in Computer and Information Science, Lyon, France, 25–27 August 2020. [Google Scholar]
- van der Spoel, E.; Rozing, M.P.; Houwing-Duistermaat, J.J.; Eline Slagboom, P.; Beekman, M.; de Craen, A.J.M.; Westendorp, R.G.J.; van Heemst, D. Siamese Neural Networks for One-Shot Image Recognition. ICML-Deep Learn. Workshop 2015. [Google Scholar] [CrossRef]
- Bromley, J.; Bentz, J.W.; Bottou, L.; Guyon, I.; Lecun, Y.; Moore, C.; Säckinger, E.; Shah, R. Signature Verification Using a “Siamese” Time Delay Neural Network. Int. J. Pattern Recognit. Artif. Intell. 1993, 7, 669–688. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Roy, S.; Harandi, M.; Nock, R.; Hartley, R. Siamese networks: The tale of two manifolds. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019; Volume 2019-Octob, pp. 3046–3055. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Z.; Zhang, T.; Wang, R. Noniterative deep learning: Incorporating restricted boltzmann machine into multilayer random weight neural networks. IEEE Trans. Syst. Man Cybern. Syst. 2019, 49, 1299–1308. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.W.; Snead, D.R.J.; Cree, I.A.; Rajpoot, N.M. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengupta, S.; Basak, S.; Saikia, P.; Paul, S.; Tsalavoutis, V.; Atiah, F.; Ravi, V.; Peters, A. A review of deep learning with special emphasis on architectures, applications and recent trends. Knowl.-Based Syst. 2020, 194. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Sulikowski, P.; Zdziebko, T. Deep learning-enhanced framework for performance evaluation of a recommending interface with varied recommendation position and intensity based on eye-tracking equipment data processing. Electronics 2020, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, K.; Huynh, N.T.; Nguyen, P.C.; Nguyen, K.D.; Vo, N.D.; Nguyen, T.V. Detecting objects from space: An evaluation of deep-learning modern approaches. Electronics 2020, 9, 583. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Lei, Y.; Li, X.; Zhang, H. Deep learning for fingerprint localization in indoor and outdoor environments. ISPRS Int. J. Geo-Inf. 2020, 9, 267. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015-Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D.D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- George, N.I.; Lu, T.-P.; Chang, C.-W. Cost-sensitive performance metric for comparing multiple ordinal classifiers. Artif. Intell. Res. 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Baccianella, S.; Esuli, A.; Sebastiani, F. Evaluation measures for ordinal regression. In Proceedings of the ISDA 2009-9th International Conference on Intelligent Systems Design and Applications, Pisa, Italy, 30 November–2 December 2009; pp. 283–287. [Google Scholar]
- Gorodkin, J. Comparing two K-category assignments by a K-category correlation coefficient. Comput. Biol. Chem. 2004, 28, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef] [PubMed]












| Layer (Type) | Output Shape | No. of Parameters |
|---|---|---|
| input_1 (InputLayer) | (None, 150, 150, 3) | 0 |
| block1_conv1 (Conv2D) | (None, 150, 150, 64) | 1792 |
| block1_conv2 (Conv2D) | (None, 150, 150, 64) | 36,928 |
| block1_pool (MaxPooling2D) | (None, 75, 75, 64) | 0 |
| dense_1 (Dense) | (None, 75, 75, 128) | 73,856 |
| block2_conv2 (Conv2D) | (None, 75, 75, 128) | 147,584 |
| block2_pool (MaxPooling2D) | (None, 37, 37, 128) | 0 |
| block3_conv1 (Conv2D) | (None, 37, 37, 256) | 295,168 |
| block3_conv2 (Conv2D) | (None, 37, 37, 256) | 590,080 |
| block3_conv3 (Conv2D) | (None, 37, 37, 256) | 590,080 |
| block3_pool (MaxPooling2D) | (None, 18, 18, 256) | 0 |
| block4_conv1 (Conv2D) | (None, 18, 18, 512) | 1,180,160 |
| block4_conv2 (Conv2D) | (None, 18, 18, 512) | 2,359,808 |
| block4_conv3 (Conv2D) | (None, 18, 18, 512) | 2,359,808 |
| block4_pool (MaxPooling2D) | (None, 9, 9, 512) | 0 |
| block5_conv1 (Conv2D) | (None, 9, 9, 512) | 2,359,808 |
| block5_conv2 (Conv2D) | (None, 9, 9, 512) | 2,359,808 |
| block5_conv3 (Conv2D) | (None, 9, 9, 512) | 2,359,808 |
| block5_pool (MaxPooling2D) | (None, 4, 4, 512) | 0 |
| flatten_1 (Flatten) | (None, 8192) | 0 |
| batch_normalization_1 (BatchNormalization) | (None, 8192) | 32,768 |
| dense_1 (Dense) | (None, 20) | 163,860 |
| dense_2 (Dense) | (None, 10) | 210 |
| Total params: 14,911,526 Trainable params: 7,259,878 Non-trainable params: 7,651,648 | ||
| Method | Cases from ISLES 2017 | MAEM |
|---|---|---|
| Random Forest for Stroke Lesion and Clinical Outcome prediction [22] | 43/43 | 1.24 |
| PMFE-SN | 43/43 | 1.18 |
| Method | Cases from ISLES 2017 | MCC | |||
|---|---|---|---|---|---|
| Random Forest for Stroke Lesion and Clinical Outcome prediction [22] | 43/43 | 0.152 | 0.21 | 0.18 | 0.04 |
| PMFE-SN | 43/43 | 0.258 | 0.31 | 0.28 | 0.09 |
| Class | ||||||
|---|---|---|---|---|---|---|
| Method | 0 | 1 | 2 | 3 | 4 | |
| State-of-the-art | Precision | 0 | 0.50 | 0.26 | 0 | 0 |
| Recall | 0 | 0.33 | 0.73 | 0 | 0 | |
| AUC | 0.50 | 0.55 | 0.50 | 0.50 | 0.50 | |
| Accuracy | 0 | 0.33 | 0.73 | 0 | 0 | |
| PMFE-SN | Precision | 0 | 0.46 | 0.33 | 0 | 0.50 |
| Recall | 0 | 0.61 | 0.27 | 0 | 0.67 | |
| AUC | 0.46 | 0.55 | 0.54 | 0.46 | 0.81 | |
| Accuracy | 0 | 0.61 | 0.27 | 0 | 0.67 | |
| PMFE-SN | MAEM | MCC | |||
|---|---|---|---|---|---|
| With data augmentation | 1.18 | 0.258 | 0.31 | 0.28 | 0.09 |
| Without data augmentation | 1.45 | 0.162 | 0.21 | 0.18 | 0.07 |
| Accuracy Per Class (PMFE-SN) | |||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| Without Augmentation | 0 | 0.33 | 0.73 | 0 | 0 |
| With augmentation | 0 | 0.61 | 0.27 | 0 | 0.67 |
| Method | Cases from ISLES 2016 | MAE |
|---|---|---|
| 1. Prediction of ischemic stroke lesion and clinical outcome in multi-modal MRI images using random forests [31] | 19/19 (test set) | 1.26 ± 0.87 |
| 2. Ensemble of deep convolutional neural networks for prognosis of ischemic stroke [33] | 19/19 (test set) | 1.10 ± 0.70 |
| 3. Predicting stroke lesion and clinical outcome with random forests [34] | 19/19 (test set) | 1.05 ± 0.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osama, S.; Zafar, K.; Sadiq, M.U. Predicting Clinical Outcome in Acute Ischemic Stroke Using Parallel Multi-Parametric Feature Embedded Siamese Network. Diagnostics 2020, 10, 858. https://doi.org/10.3390/diagnostics10110858
Osama S, Zafar K, Sadiq MU. Predicting Clinical Outcome in Acute Ischemic Stroke Using Parallel Multi-Parametric Feature Embedded Siamese Network. Diagnostics. 2020; 10(11):858. https://doi.org/10.3390/diagnostics10110858
Chicago/Turabian StyleOsama, Saira, Kashif Zafar, and Muhammad Usman Sadiq. 2020. "Predicting Clinical Outcome in Acute Ischemic Stroke Using Parallel Multi-Parametric Feature Embedded Siamese Network" Diagnostics 10, no. 11: 858. https://doi.org/10.3390/diagnostics10110858






