CoSinGAN: Learning COVID-19 Infection Segmentation from a Single Radiological Image
Abstract
:1. Introduction
2. Material and Methodology
2.1. Methodology
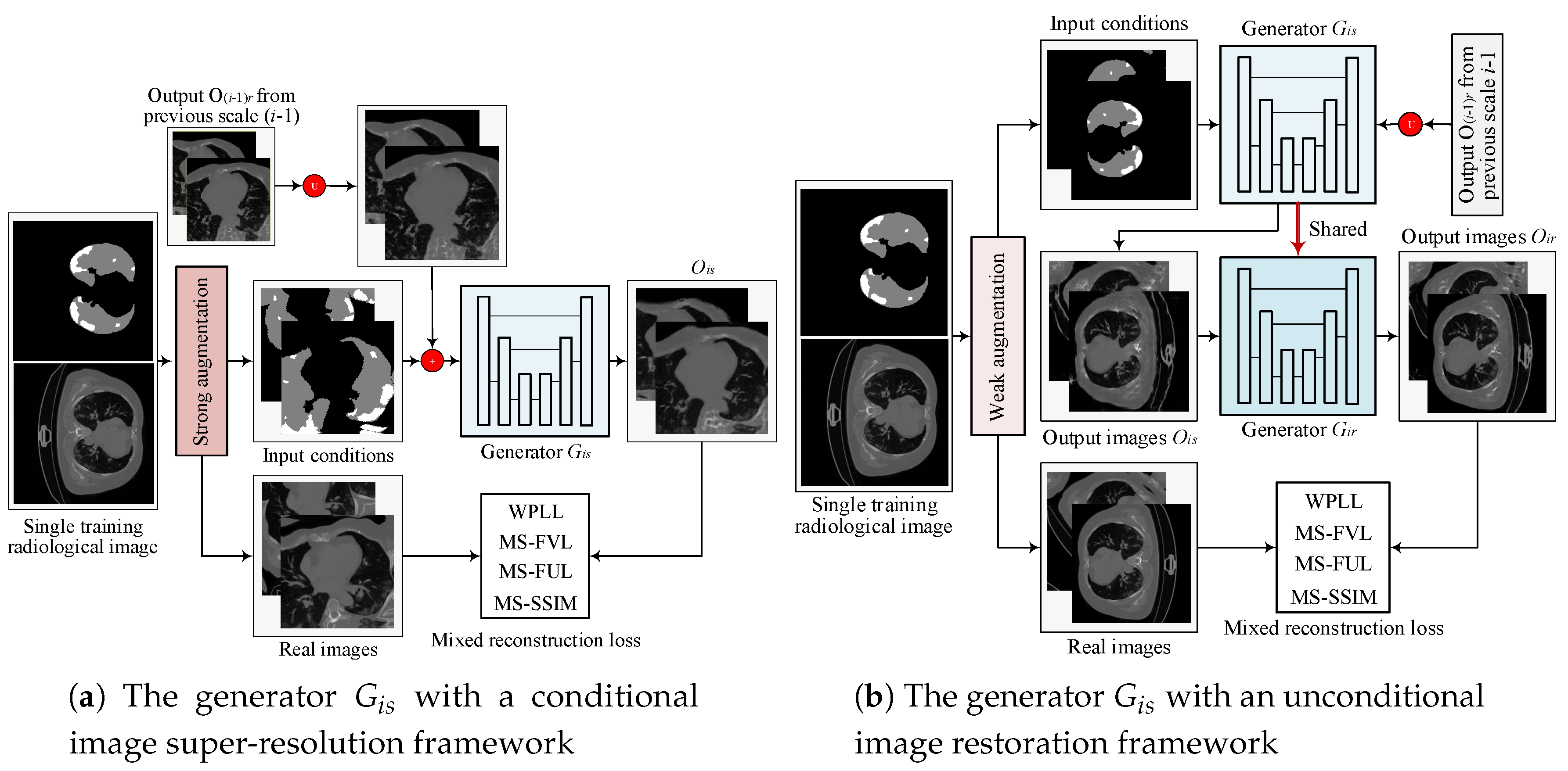
2.1.1. Multi-Scale Architecture with a Pyramid of Two-Stage GANs
2.1.2. Objective
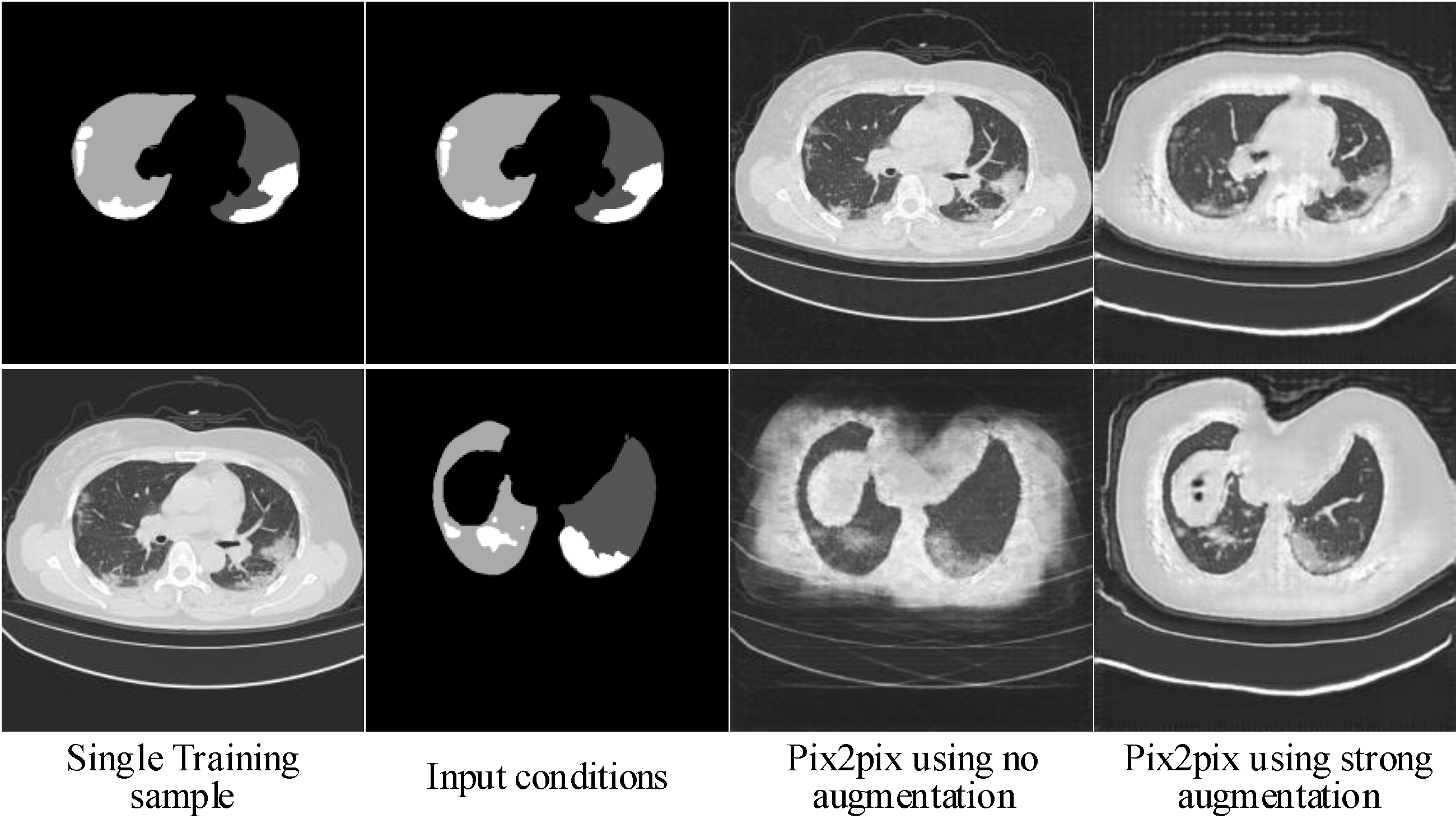
2.1.3. Hierarchical Data Augmentation
- SA is critical for to generalize to different input conditions.
- WA helps to fit the real image distribution without introducing additional learning burden.
- Decreasing the intensity of SA along with the increasing of image scales can handle the balance between fitting conditions and fitting images well.
2.2. Materials
3. Experiment Results and Discussion
3.1. Experiments on Synthesizing Radiological Images
3.1.1. Training Details
3.1.2. Ablation Experiments
3.1.3. Evaluation and Comparison on Image Quality
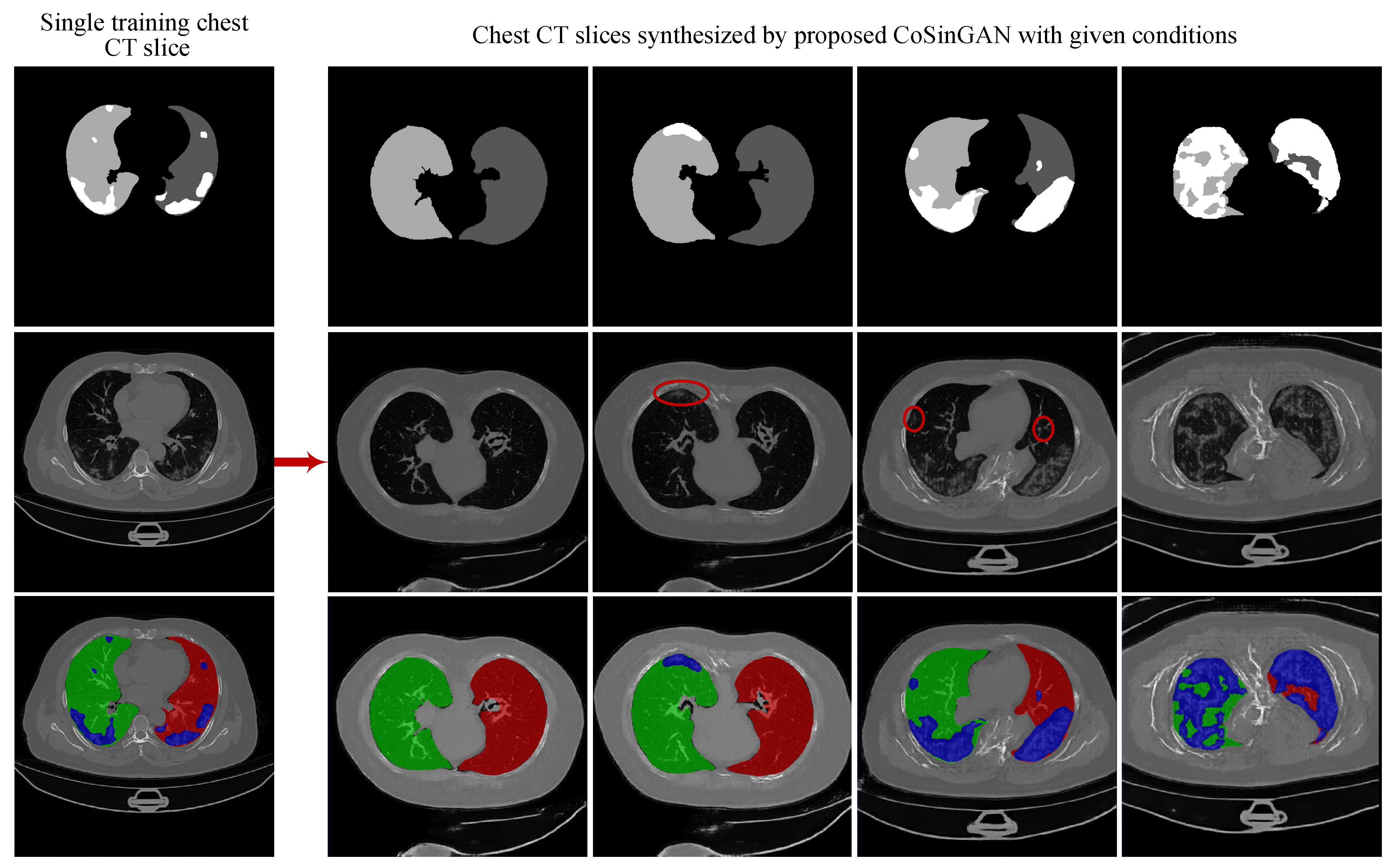
3.1.4. Evaluation on the Ability of CoSinGAN in Generating Diverse Samples
3.2. Experiments on Learning Deep Models for Automated Lung and Infection Segmentation
3.2.1. Baselines
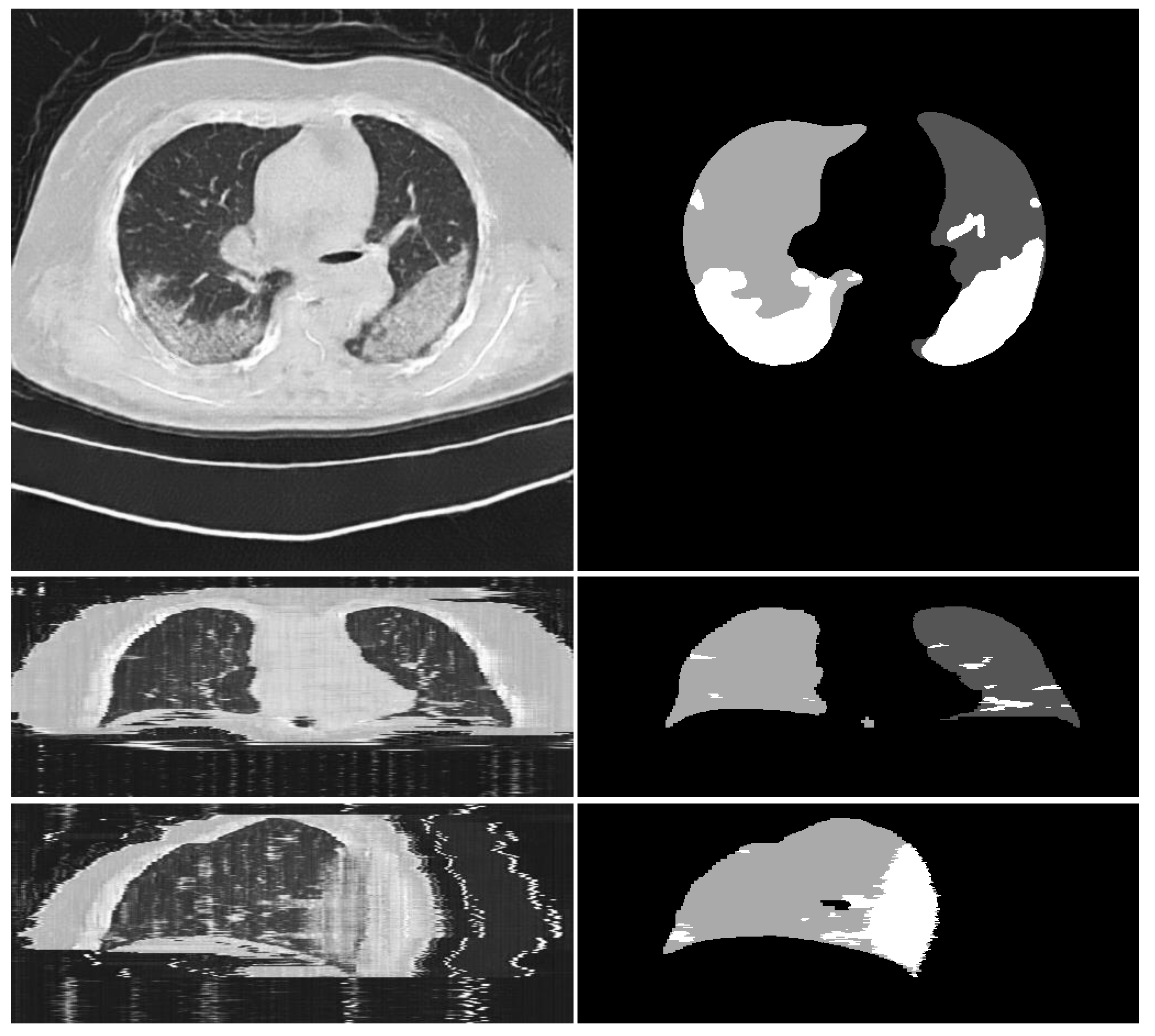
3.2.2. CoSinGAN, IF-CoSinGAN, and RC-CoSinGAN
3.2.3. Segmentation Models and Training Details
3.2.4. Evaluation Metrics
3.2.5. Results and Discussion
- (1)
- Quantitative comparison
- (2)
- Qualitative comparison
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020, 20, 425–434. [Google Scholar] [CrossRef]
- WHO Coronavirus Disease (COVID-19) Dashboard. 2020. Available online: https://covid19.who.int (accessed on 8 October 2020).
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Xia, L. Coronavirus disease 2019 (COVID-19): Role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020, 214, 1280–1286. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020. [Google Scholar] [CrossRef]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev. Biomed. Eng. 2020. [Google Scholar] [CrossRef] [Green Version]
- Mei, X.; Lee, H.C.; Diao, K.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence-enabled rapid diagnosis of COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Kanne, J.P. Chest CT Findings in 2019 Novel Coronavirus (2019-nCoV) Infections from Wuhan, China: Key Points for the Radiologist. Radiology 2020, 295, 16–17. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Xia, L.; Yan, F.; Wan, Z.; Shi, F.; Yuan, H.; Jiang, H.; Wu, D.; Sui, H.; Zhang, C.; et al. Diagnosis of coronavirus disease 2019 (covid-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging 2020, 39, 2606–2614. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wang, B.; Gong, D.; Luo, C.; Zhao, W.; Shen, J.; Shi, Q.; Jin, S.; Zhang, L.; You, Z. COVID-19 Chest CT Image Segmentation–A Deep Convolutional Neural Network Solution. arXiv 2020, arXiv:2004.10987. [Google Scholar]
- Ouyang, X.; Huo, J.; Xia, L.; Shan, F.; Liu, J.; Mo, Z.; Yan, F.; Ding, Z.; Yang, Q.; Song, B.; et al. Dual-Sampling Attention Network for Diagnosis of COVID-19 from Community Acquired Pneumonia. arXiv 2020, arXiv:2005.02690. [Google Scholar]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loey, M.; Smarandache, F.; M Khalifa, N.E. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef] [Green Version]
- Waheed, A.; Goyal, M.; Gupta, D.; Khanna, A.; Al-Turjman, F.; Pinheiro, P.R. CovidGAN: Data Augmentation Using Auxiliary Classifier GAN for Improved Covid-19 Detection. IEEE Access 2020, 8, 91916–91923. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.; An, X.; Ge, C.; Yu, Z.; Chen, J.; Zhu, Q.; Dong, G.; He, J.; He, Z.; et al. Towards Efficient COVID-19 CT Annotation: A Benchmark for Lung and Infection Segmentation. arXiv 2020, arXiv:2004.12537. [Google Scholar]
- Karras, T.; Laine, S.; Aittala, M.; Hellsten, J.; Lehtinen, J.; Aila, T. Analyzing and improving the image quality of stylegan. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 8110–8119. [Google Scholar]
- Hinz, T.; Heinrich, S.; Wermter, S. Generating multiple objects at spatially distinct locations. arXiv 2019, arXiv:1901.00686. [Google Scholar]
- Karras, T.; Laine, S.; Aila, T. A style-based generator architecture for generative adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, CA, USA, 18–20 June 2019; pp. 4401–4410. [Google Scholar]
- Wang, X.; Yu, K.; Wu, S.; Gu, J.; Liu, Y.; Dong, C.; Qiao, Y.; Change Loy, C. Esrgan: Enhanced super-resolution generative adversarial networks. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Yu, X.; Qu, Y.; Hong, M. Underwater-GAN: Underwater image restoration via conditional generative adversarial network. In Proceedings of the International Conference on Pattern Recognition, Beijing, China, 20–24 August 2018; pp. 66–75. [Google Scholar]
- Bau, D.; Strobelt, H.; Peebles, W.; Zhou, B.; Zhu, J.Y.; Torralba, A. Semantic photo manipulation with a generative image prior. arXiv 2020, arXiv:2005.07727. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Hinz, T.; Fisher, M.; Wang, O.; Wermter, S. Improved Techniques for Training Single-Image GANs. arXiv 2020, arXiv:2003.11512. [Google Scholar]
- Zhou, Y.; Zhu, Z.; Bai, X.; Lischinski, D.; Cohen-Or, D.; Huang, H. Non-stationary texture synthesis by adversarial expansion. arXiv 2018, arXiv:1805.04487. [Google Scholar] [CrossRef] [Green Version]
- Shocher, A.; Bagon, S.; Isola, P.; Irani, M. InGAN: Capturing and Remapping the “DNA” of a Natural Image. arXiv 2018, arXiv:1812.00231. [Google Scholar]
- Shaham, T.R.; Dekel, T.; Michaeli, T. Singan: Learning a generative model from a single natural image. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 4570–4580. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Isensee, F.; Petersen, J.; Kohl, S.A.; Jäger, P.F.; Maier-Hein, K.H. nnu-net: Breaking the spell on successful medical image segmentation. arXiv 2019, arXiv:1904.08128. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Wang, Z.; Simoncelli, E.P.; Bovik, A.C. Multiscale structural similarity for image quality assessment. In Proceedings of the Thrity-Seventh Asilomar Conference on Signals, Systems & Computers, Pacific Grove, CA, USA, 9–12 November 2003; Volume 2, pp. 1398–1402. [Google Scholar]
- Zhao, H.; Gallo, O.; Frosio, I.; Kautz, J. Loss functions for image restoration with neural networks. IEEE Trans. Comput. Imaging 2016, 3, 47–57. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [Green Version]
- Dosovitskiy, A.; Brox, T. Generating images with perceptual similarity metrics based on deep networks. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 658–666. [Google Scholar]
- Johnson, J.; Alahi, A.; Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 10–16 October 2016; pp. 694–711. [Google Scholar]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-resolution image synthesis and semantic manipulation with conditional gans. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8798–8807. [Google Scholar]
- Morozov, S.; Andreychenko, A.; Pavlov, N.; Vladzymyrskyy, A.; Ledikhova, N.; Gombolevskiy, V.; Blokhin, I.; Gelezhe, P.; Gonchar, A.; Chernina, V.; et al. MosMedData: Chest CT Scans with COVID-19 Related Findings. medRxiv 2020. [Google Scholar] [CrossRef]
- Kiser, K.; Barman, A.; Stieb, S.; Fuller, C.D.; Giancardo, L. Novel Autosegmentation Spatial Similarity Metrics Capture the Time Required to Correct Segmentations Better than Traditional Metrics in a Thoracic Cavity Segmentation Workflow. medRxiv 2020. [Google Scholar] [CrossRef]
- Nikolov, S.; Blackwell, S.; Mendes, R.; De Fauw, J.; Meyer, C.; Hughes, C.; Askham, H.; Romera-Paredes, B.; Karthikesalingam, A.; Chu, C.; et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. arXiv 2018, arXiv:1809.04430. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Automatic Structure Segmentation for Radiotherapy Planning Challenge. 2019. Available online: https://structseg2019.grand-challenge.org (accessed on 1 July 2020).












| Subtask | Model | Left Lung | Right Lung | Infection (COVID-19-CT-Seg) | |||
|---|---|---|---|---|---|---|---|
| DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | ||
| Fold-0 | Pix2pix | 91.4 ± 6.8 | 72.7 ± 11.7 | 91.7 ± 6.3 | 69.1 ± 11.4 | 46.3 ± 19.8 | 43.8 ± 20.1 |
| Enhanced pix2pix | 86.9 ± 15.4 | 69.1 ± 16.3 | 88.9 ± 11.4 | 68.7 ± 15.8 | 50.4 ± 22.6 | 47.3 ± 21.1 | |
| CoSinGAN | 93.1 ± 6.3 | 76.7 ± 11.5 | 94.5 ± 4.1 | 76.4 ± 10.3 | 37.0 ± 21.0 | 35.6 ± 20.8 | |
| Fold-1 | Pix2pix | 86.0 ± 10.3 | 60.0 ± 15.3 | 87.2 ± 10.8 | 60.2 ± 15.3 | 42.7 ± 22.7 | 36.5 ± 23.8 |
| Enhanced pix2pix | 92.6 ± 5.9 | 74.6 ± 10.5 | 94.2 ± 3.6 | 75.3 ± 8.3 | 60.3 ± 20.3 | 55.9 ± 24.0 | |
| CoSinGAN | 91.9 ± 7.0 | 74.0 ± 12.1 | 94.2 ± 3.9 | 74.6 ± 9.6 | 60.3 ± 21.1 | 53.8 ± 23.4 | |
| Fold-2 | Pix2pix | 81.6 ± 11.7 | 52.2 ± 19.2 | 80.5 ± 13.4 | 49.7 ± 17.8 | 41.2 ± 16.6 | 39.7 ± 18.0 |
| Enhanced pix2pix | 92.5 ± 4.5 | 70.7 ± 9.7 | 93.4 ± 3.3 | 70.1 ± 7.3 | 44.0 ± 20.8 | 43.8 ± 22.5 | |
| CoSinGAN | 91.9 ± 6.4 | 73.0 ± 11.2 | 92.5 ± 4.8 | 70.6 ± 8.9 | 48.1 ± 19.6 | 45.0 ± 20.3 | |
| Fold-3 | Pix2pix | 83.4 ± 18.6 | 57.3 ± 18.5 | 83.7 ± 17.6 | 54.3 ± 19.6 | 40.3 ± 22.0 | 38.7 ± 23.2 |
| Enhanced pix2pix | 92.5 ± 7.0 | 73.0 ± 12.1 | 93.4 ± 4.0 | 69.8 ± 10.4 | 55.1 ± 19.3 | 50.6 ± 23.4 | |
| CoSinGAN | 94.5 ± 5.2 | 79.8 ± 9.9 | 95.3 ± 2.5 | 78.3 ± 8.6 | 62.6 ± 20.0 | 59.3 ± 23.0 | |
| Fold-4 | Pix2pix | 86.2 ± 13.1 | 59.6 ± 13.7 | 87.4 ± 9.8 | 57.9 ± 13.1 | 43.1 ± 14.9 | 41.1 ± 16.0 |
| Enhanced pix2pix | 90.1 ± 9.7 | 69.3 ± 14.0 | 91.0 ± 6.3 | 67.1 ± 13.6 | 48.3 ± 13.9 | 49.2 ± 18.2 | |
| CoSinGAN | 91.8 ± 5.6 | 71.4 ± 10.3 | 92.3 ± 6.0 | 69.8 ± 11.0 | 62.4 ± 12.6 | 58.5 ± 15.9 | |
| Avg | Pix2pix | 85.8 ± 13.1 | 60.3 ± 17.3 | 86.1 ± 12.7 | 58.2 ± 17.0 | 42.7 ± 19.5 | 40.0 ± 20.6 |
| Enhanced pix2pix | 90.9 ± 9.6 | 71.3 ± 12.9 | 92.2 ± 6.8 | 70.2 ± 11.9 | 51.6 ± 20.4 | 49.4 ± 22.2 | |
| CoSinGAN | 92.6 ± 6.2 | 75.0 ± 11.4 | 93.8 ± 4.6 | 73.9 ± 10.3 | 54.1 ± 21.6 | 50.4 ± 22.7 | |
| Methods | COVID-19-CT-Seg Dataset (20 CT Cases) | MosMed Dataset (50 CT Cases) | |
|---|---|---|---|
| Training | Testing | Testing | |
| Strong baselines (Benchmark) | 4 CT cases with an averageof 704 CT slices | 16 CT cases | 50 CT cases |
| CoSinGAN | 4 CT slices → 80 synthetic CT volumes | 16 CT cases | 50 CT cases |
| IF-CoSinGAN | 4 CT slices → 80 synthetic CT volumes | 16 CT cases | 50 CT cases |
| RC-CoSinGAN | 4 CT slices → 80 synthetic CT volumes | 16 CT cases | 50 CT cases |
| Methods | Training | Testing |
|---|---|---|
| Task2-MSD | MSD Lung Tumor dataset (51 CT cases) | COVID-19-CT-Seg dataset (20 CT cases) |
| Task2-StructSeg | StructSeg2019 (40 CT cases) | COVID-19-CT-Seg dataset (20 CT cases) |
| Task2-NSCLC | NSCLC Pleural Effusion dataset (62 CT cases) | COVID-19-CT-Seg dataset (20 CT cases) |
| Methods | Training | Testing | |
|---|---|---|---|
| Task3-MSD | MSD Lung Tumor dataset (51 CT cases) | COVID-19-CT-Seg dataset (4 CT cases) | COVID-19-CT-Seg dataset (16 CT cases) |
| Task3-StructSeg | StructSeg2019 (40 CT cases) | COVID-19-CT-Seg dataset (4 CT cases) | COVID-19-CT-Seg dataset (16 CT cases) |
| Task3-NSCLC | NSCLC Pleural Effusion dataset (62 CT cases) | COVID-19-CT-Seg dataset (4 CT cases) | COVID-19-CT-Seg dataset (16 CT cases) |
| Subtask | Methods | Slice Number | Left Lung | Right Lung | Infection (COVID-19-CT-Seg) | Infection (MosMed) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | |||
| Fold-0 | Benchmark | 634 | 89.9 ± 10.7 | 72.8 ± 18.6 | 91.3 ± 8.6 | 73.5 ± 18.1 | 67.8 ± 20.7 | 69.1 ± 23.0 | 53.2 ± 20.1 | 62.5 ± 18.7 |
| CoSinGAN | 4 | 92.6 ± 7.9 | 78.4 ± 11.9 | 93.9 ± 5.1 | 77.5 ± 10.1 | 61.9 ± 19.3 | 55.6 ± 21.2 | 39.5 ± 24.6 | 47.5 ± 23.6 | |
| IF-CoSinGAN | 4 | 93.8 ± 5.2 | 79.0 ± 9.7 | 94.7 ± 3.8 | 78.3 ± 9.5 | 58.7 ± 20.5 | 54.2 ± 20.4 | 29.0 ± 20.1 | 38.1 ± 19.6 | |
| RC-CoSinGAN | 4 | 94.1 ± 5.5 | 81.8 ± 10.3 | 95.0 ± 3.8 | 80.2 ± 8.8 | 73.9 ± 20.2 | 73.7 ± 21.8 | 54.0 ± 22.4 | 62.0 ± 21.3 | |
| Fold-1 | Benchmark | 681 | 87.7 ± 13.9 | 68.6 ± 17.5 | 90.4 ± 9.3 | 70.4 ± 14.4 | 61.5 ± 21.6 | 58.1 ± 26.8 | 44.5 ± 19.9 | 51.4 ± 19.6 |
| CoSinGAN | 4 | 93.4 ± 4.6 | 75.9 ± 8.6 | 94.3 ± 3.2 | 75.6 ± 8.6 | 67.9 ± 19.6 | 62.8 ± 21.8 | 52.0 ± 21.3 | 59.6 ± 20.2 | |
| IF-CoSinGAN | 4 | 93.7 ± 4.9 | 77.1 ± 9.4 | 94.7 ± 3.4 | 76.9 ± 8.0 | 67.1 ± 19.7 | 61.1 ± 22.2 | 41.8 ± 21.7 | 48.6 ± 19.9 | |
| RC-CoSinGAN | 4 | 93.6 ± 4.7 | 77.8 ± 8.6 | 94.6 ± 3.2 | 77.1 ± 7.4 | 71.3 ± 20.4 | 70.5 ± 22.5 | 41.9 ± 22.5 | 49.5 ± 22.7 | |
| Fold-2 | Benchmark | 683 | 91.1 ± 12.4 | 76.0 ± 16.1 | 92.5 ± 8.8 | 74.2 ± 15.6 | 52.5 ± 25.2 | 49.1 ± 25.0 | 41.2 ± 22.6 | 45.2 ± 21.4 |
| CoSinGAN | 4 | 93.3 ± 5.2 | 76.2 ± 9.4 | 94.2 ± 3.7 | 75.1 ± 8.2 | 61.7 ± 21.4 | 59.3 ± 23.0 | 36.4 ± 22.0 | 43.2 ± 20.8 | |
| IF-CoSinGAN | 4 | 93.1 ± 5.3 | 75.5 ± 10.4 | 93.9 ± 4.1 | 74.0 ± 9.4 | 60.8 ± 19.7 | 56.7 ± 21.9 | 31.6 ± 22.1 | 38.4 ± 19.9 | |
| RC-CoSinGAN | 4 | 94.2 ± 5.0 | 79.3 ± 9.6 | 94.6 ± 4.5 | 77.7 ± 9.6 | 73.3 ± 19.8 | 74.9 ± 21.3 | 52.1 ± 20.4 | 59.3 ± 19.8 | |
| Fold-3 | Benchmark | 649 | 78.1 ± 22.4 | 60.9 ± 20.2 | 80.7 ± 19.8 | 62.0 ± 19.9 | 57.9 ± 27.6 | 57.8 ± 31.8 | 42.0 ± 24.1 | 48.9 ± 25.4 |
| CoSinGAN | 4 | 93.5 ± 4.9 | 76.4 ± 9.1 | 94.7 ± 2.1 | 76.7 ± 7.7 | 63.5 ± 19.2 | 62.8 ± 22.7 | 51.0 ± 22.9 | 59.5 ± 21.4 | |
| IF-CoSinGAN | 4 | 93.2 ± 7.1 | 77.7 ± 10.5 | 94.6 ± 3.7 | 77.2 ± 9.3 | 66.7 ± 19.7 | 66.5 ± 23.2 | 50.5 ± 21.5 | 58.8 ± 20.0 | |
| RC-CoSinGAN | 4 | 94.1 ± 7.1 | 81.0 ± 10.1 | 95.1 ± 3.6 | 79.7 ± 8.5 | 64.8 ± 21.6 | 67.4 ± 23.0 | 46.6 ± 24.2 | 54.2 ± 22.5 | |
| Fold-4 | Benchmark | 873 | 89.9 ± 12.4 | 74.0 ± 16.2 | 92.0 ± 8.7 | 74.8 ± 16.0 | 48.6 ± 29.8 | 52.0 ± 32.1 | 22.8 ± 22.9 | 25.6 ± 22.4 |
| CoSinGAN | 4 | 92.2 ± 6.9 | 75.5 ± 10.0 | 93.2 ± 4.8 | 73.9 ± 10.2 | 65.4 ± 11.7 | 62.2 ± 15.8 | 32.8 ± 24.6 | 40.4 ± 22.9 | |
| IF-CoSinGAN | 4 | 92.4 ± 6.4 | 74.6 ± 9.6 | 93.4 ± 4.0 | 72.6 ± 9.0 | 70.1 ± 9.3 | 68.5 ± 13.5 | 32.6 ± 23.8 | 40.9 ± 22.9 | |
| RC-CoSinGAN | 4 | 93.4 ± 5.2 | 77.3 ± 8.7 | 93.9 ± 3.7 | 75.6 ± 9.1 | 73.3 ± 8.0 | 73.5 ± 13.6 | 42.3 ± 26.0 | 49.1 ± 26.6 | |
| Avg | Benchmark | 704 | 87.3 ± 15.7 | 70.5 ± 18.6 | 89.4 ± 12.7 | 71.0 ± 17.6 | 57.7 ± 26.1 | 57.2 ± 28.8 | 40.7 ± 24.1 | 46.7 ± 24.7 |
| CoSinGAN | 4 | 93.0 ± 6.0 | 76.5 ± 11.4 | 94.1 ± 4.0 | 75.8 ± 9.1 | 64.1 ± 18.7 | 60.5 ± 21.3 | 42.4 ± 24.4 | 50.0 ± 23.3 | |
| IF-CoSinGAN | 4 | 93.2 ± 5.9 | 76.8 ± 10.1 | 94.3 ± 3.8 | 75.8 ± 9.3 | 64.7 ± 18.8 | 61.4 ± 21.3 | 37.1 ± 23.4 | 45.0 ± 22.0 | |
| RC-CoSinGAN | 4 | 93.9 ± 5.6 | 79.5 ± 9.6 | 94.6 ± 3.8 | 78.1 ± 8.9 | 71.3 ± 19.0 | 72.0 ± 20.9 | 47.4 ± 23.7 | 54.8 ± 23.3 | |
| Subtask | Methods | Slice Number | Left Lung | Right Lung | Infection (COVID-19-CT-Seg) | Infection (MosMed) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | DSC ↑ | NSD ↑ | |||
| Fold-0 | Benchmark | 634 | 53.8 ± 28.4 | 39.1 ± 18.3 | 65.5 ± 19.4 | 47.4 ± 14.3 | 65.4 ± 23.9 | 68.2 ± 23.2 | 51.0 ± 23.2 | 60.1 ± 22.4 |
| CoSinGAN | 4 | 59.9 ± 9.8 | 40.1 ± 6.6 | 56.6 ± 14.2 | 32.6 ± 6.9 | 51.7 ± 21.1 | 52.3 ± 21.9 | 37.7 ± 24.0 | 45.5 ± 23.8 | |
| Fold-1 | Benchmark | 681 | 40.3 ± 18.7 | 27.5 ± 12.0 | 60.1 ± 11.1 | 41.7 ± 9.9 | 64.7 ± 21.8 | 60.6 ± 25.1 | 49.8 ± 21.3 | 55.4 ± 20.1 |
| CoSinGAN | 4 | 60.7 ± 10.2 | 36.9 ± 6.7 | 61.7 ± 8.7 | 34.2 ± 6.2 | 67.8 ± 19.5 | 63.7 ± 22.1 | 52.8 ± 23.7 | 59.3 ± 23.4 | |
| Fold-2 | Benchmark | 683 | 80.3 ± 18.8 | 66.8 ± 18.8 | 85.2 ± 12.4 | 68.6 ± 15.1 | 60.7 ± 27.6 | 62.5 ± 28.9 | 46.8 ± 24.2 | 50.2 ± 24.2 |
| CoSinGAN | 4 | 59.9 ± 10.9 | 40.5 ± 6.4 | 56.0 ± 14.7 | 30.8 ± 8.9 | 64.2 ± 19.4 | 64.4 ± 20.9 | 39.7 ± 23.4 | 47.0 ± 23.0 | |
| Fold-3 | Benchmark | 649 | 79.7 ± 13.6 | 65.4 ± 14.4 | 84.0 ± 9.8 | 67.7 ± 13.0 | 62.0 ± 27.9 | 65.3 ± 28.9 | 44.8 ± 26.6 | 52.7 ± 25.4 |
| CoSinGAN | 4 | 59.5 ± 7.1 | 41.6 ± 5.1 | 44.5 ± 15.6 | 24.4 ± 8.2 | 59.9 ± 21.1 | 60.5 ± 24.1 | 51.0 ± 23.5 | 57.0 ± 22.4 | |
| Fold-4 | Benchmark | 873 | 72.4 ± 21.1 | 58.6 ± 20.8 | 80.9 ± 13.4 | 63.4 ± 15.9 | 51.4 ± 30.2 | 51.9 ± 31.0 | 16.7 ± 19.5 | 19.3 ± 19.0 |
| CoSinGAN | 4 | 57.4 ± 13.4 | 37.6 ± 7.3 | 52.6 ± 16.4 | 33.3 ± 7.5 | 64.1 ± 15.4 | 63.2 ± 17.0 | 43.4 ± 22.8 | 49.8 ± 22.0 | |
| Avg | Benchmark | 704 | 65.4 ± 25.6 | 51.6 ± 22.9 | 75.3 ± 16.8 | 57.9 ± 17.5 | 60.8 ± 26.3 | 61.6 ± 27.5 | 41.8 ± 26.4 | 47.6 ± 26.7 |
| CoSinGAN | 4 | 59.5 ± 10.5 | 39.3 ± 6.7 | 54.3 ± 15.3 | 31.2 ± 8.4 | 61.5 ± 20.2 | 60.8 ± 21.8 | 44.9 ± 24.2 | 51.7 ± 23.6 | |
| Methods | Infection (COVID-19-CT-Seg) | ||
|---|---|---|---|
| DSC ↑ | NSD ↑ | ||
| Learning with only non-COVID-19 CT scans | Task2-MSD | 7.9 ± 11.5 | 12.9 ± 15.3 |
| Task2-StructSeg | 0.2 ± 0.8 | 0.6 ± 1.6 | |
| Task2-NSCLC | 1.2 ± 2.9 | 7.3 ± 9.7 | |
| Learning with both COVID-19 and non-COVID-19 CT scans | Task3-MSD | 51.2 ± 26.8 | 52.7 ± 27.4 |
| Task3-StructSeg | 57.4 ± 26.6 | 57.3 ± 28.4 | |
| Task3-NSCLC | 52.5 ± 29.6 | 52.6 ± 30.3 | |
| Learning with only COVID-19 CT scans (or slices) | Benchmark | 57.7 ± 26.1 | 57.2 ± 28.8 |
| CoSinGAN | 64.1 ± 18.7 | 60.5 ± 21.3 | |
| RC-CoSinGAN | 71.3 ± 19.0 | 72.0 ± 20.9 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Zhong, Y.; Deng, Y.; Tang, X.; Li, X. CoSinGAN: Learning COVID-19 Infection Segmentation from a Single Radiological Image. Diagnostics 2020, 10, 901. https://doi.org/10.3390/diagnostics10110901
Zhang P, Zhong Y, Deng Y, Tang X, Li X. CoSinGAN: Learning COVID-19 Infection Segmentation from a Single Radiological Image. Diagnostics. 2020; 10(11):901. https://doi.org/10.3390/diagnostics10110901
Chicago/Turabian StyleZhang, Pengyi, Yunxin Zhong, Yulin Deng, Xiaoying Tang, and Xiaoqiong Li. 2020. "CoSinGAN: Learning COVID-19 Infection Segmentation from a Single Radiological Image" Diagnostics 10, no. 11: 901. https://doi.org/10.3390/diagnostics10110901
APA StyleZhang, P., Zhong, Y., Deng, Y., Tang, X., & Li, X. (2020). CoSinGAN: Learning COVID-19 Infection Segmentation from a Single Radiological Image. Diagnostics, 10(11), 901. https://doi.org/10.3390/diagnostics10110901






