Development of Diagnostic Tests for Detection of SARS-CoV-2
Abstract
:1. Introduction
2. Components of SARS-CoV-2 Used for Detection
2.1. Non-Structural Proteins
2.2. Structural Proteins
2.3. Antibodies
2.4. Interleukin-6
2.5. Glycans
3. Factors that Affect the Development of Diagnostic Tests for SARS-CoV-2
3.1. Materials Evaluation
3.2. Sensitivity, Specificity, and Accuracy
4. Diagnostic Tests for Detection of SARS-CoV-2
4.1. Molecular Assays for Detection of Viral Nucleic Acids
4.1.1. Polymerase Chain Reaction (PCR)
4.1.2. Isothermal Amplification
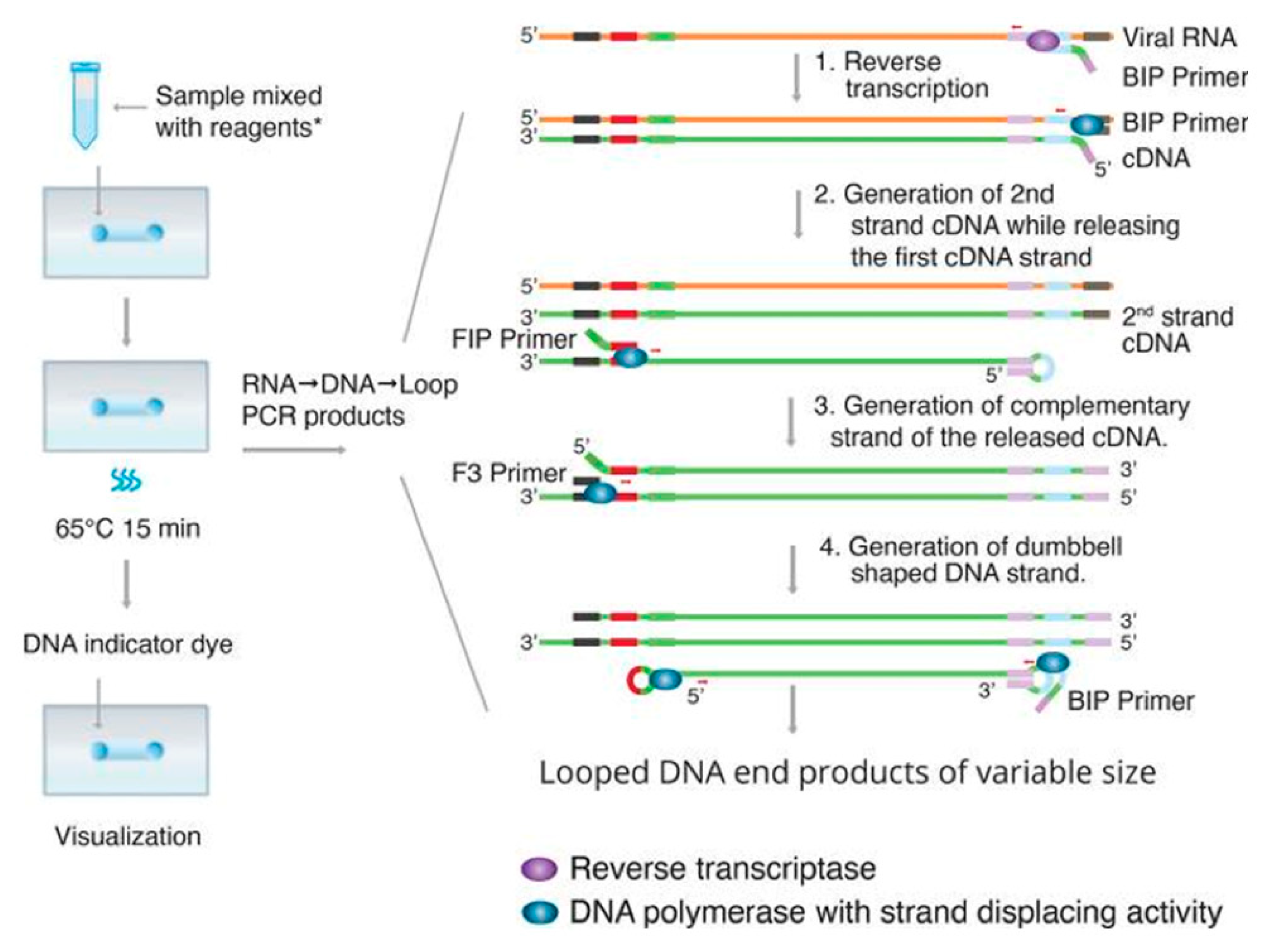
- Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP)
- Transcription—Mediated Amplification (TMA)
4.1.3. CRISPR-Based Nucleic Acid Detection
4.1.4. High-Throughput Technologies
- Next Generation Sequencing (NGS)
- Amplicon-Based Metagenomic Sequencing
4.2. Rapid Diagnostic Tests (RDT)
4.2.1. Antibody Testing
- Enzyme-Linked Immunosorbent Assay (ELISA)
- Chemiluminescence Immunoassay (CLIA)
- Lateral Flow Immunoassay (LFIA)
4.2.2. Antigen Testing
4.2.3. Proteome Peptide Microarray (PPM)
4.3. Medical Imaging
4.4. Potential Diagnostic Tests
4.4.1. Vertical Flow Assays (VFA)
4.4.2. Microfluidic Devices
5. Test Considerations
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Velavan, T.P.; Meyer, C.G. The COVID-19 epidemic. Trop. Med. Int. Health 2020, 25, 278–280. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.S.; Smith, D.W. COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don’t. Microbiol. Aust. 2020, MA20013. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, L.-L.; Wang, Y.-M.; Wu, Z.-Q.; Xiang, Z.-C.; Guo, L.; Xu, T.; Jiang, Y.-Z.; Xiong, Y.; Li, Y.-J.; Li, X.-W.; et al. Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. 2020, 133, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Dhama, K.; Sharun, K.; Iqbal Yatoo, M.; Malik, Y.S.; Singh, R.; Michalak, I.; Sah, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. COVID-19: Animals, veterinary and zoonotic links. Vet. Q. 2020, 1–22. [Google Scholar] [CrossRef]
- WHO. Modes of Transmission of Virus Causing COVID-19: Implications for IPC Precaution Recommendations. Available online: https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations (accessed on 4 November 2020).
- Worldometer. Covid-19 Coronavirus Pandemic. Available online: https://www.worldometers.info/coronavirus/ (accessed on 4 November 2020).
- CDC. United States COVID-19 Cases and Deaths by State. Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/us-cases-deaths.html (accessed on 4 November 2020).
- BLS. Civilian Unemployment Rate. Available online: https://www.bls.gov/charts/employment-situation/civilian-unemployment-rate.htm (accessed on 4 November 2020).
- Nicola, M.; Alsafi, Z.; Sohrabi, C.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, M.; Agha, R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020, 78, 185–193. [Google Scholar] [CrossRef]
- Donthu, N.; Gustafsson, A. Effects of COVID-19 on business and research. J. Bus. Res. 2020, 117, 284–289. [Google Scholar] [CrossRef]
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Rapid Commun. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolai, L.A.; Meyer, C.G.; Kremsner, P.G.; Velavan, T.P. Asymptomatic SARS Coronavirus 2 infection: Invisible yet invincible. Int. J. Infect. Dis. 2020, 100, 112–116. [Google Scholar] [CrossRef]
- Ing, A.J.; Cocks, C.; Green, J.P. COVID-19: In the footsteps of Ernest Shackleton. Thorax 2020, 75, 693–694. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- CDC. Similarities and Differences between Flu and COVID-19. Available online: https://www.cdc.gov/flu/symptoms/flu-vs-covid19.htm (accessed on 4 November 2020).
- Li, X.; Geng, M.; Peng, Y.; Meng, L.; Lu, S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020. [Google Scholar] [CrossRef]
- Chan, J.F.; Yip, C.C.; To, K.K.; Tang, T.H.; Wong, S.C.; Leung, K.H.; Fung, A.Y.; Ng, A.C.; Zou, Z.; Tsoi, H.W.; et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020, 58, e00310–e00320. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Tong, J.; Liu, M.; Shen, Y.; Guo, D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2. J. Mol. Diagn. 2020. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wu, S.; Hao, X.; Li, X.; Liu, X.; Ye, S.; Han, H.; Dong, X.; Li, X.; Li, J.; et al. Rapid colorimetric detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform: iLACO. medRxiv 2020. [Google Scholar] [CrossRef]
- Adams, E.R.; Ainsworth, M.; Anand, R.; Andersson, M.I.; Auckland, K.; Baillie, J.K.; Barnes, E.; Beer, S.; Bell, J.; Berry, T.; et al. Antibody testing for COVID-19: A report from the National COVID Scientific Advisory Panel. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, M.; Jin, R.; Peng, Y.; Wang, C.; Ren, W.; Lv, F.; Gong, S.; Fang, F.; Wang, Q.; Li, J.; et al. Generation of antibodies against Covid-19 virus for development of diagnostic tools. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Lucia, C.; Federico, P.-B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus 2 SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Hou, X.; Wu, X.; Liang, T.; Zhang, X.; Wang, D.; Teng, F.; Dai, J.; Duan, H.; Guo, S.; et al. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020. [Google Scholar] [CrossRef] [Green Version]
- Venkataraman, S.; Prasad, B.; Selvarajan, R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Li, S.Y.; Yang, X.L.; Huang, H.M.; Zhang, Y.J.; Guo, H.; Luo, C.M.; Miller, M.; Zhu, G.; Chmura, A.A.; et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018, 33, 104–107. [Google Scholar] [CrossRef] [Green Version]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Surjit, M.; Lal, S.K. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect. Genet. Evol. 2008, 8, 397–405. [Google Scholar] [CrossRef]
- Jacofsky, D.; Jacofsky, E.M.; Jacofsky, M. Understanding Antibody Testing for COVID-19. J. Arthroplasty 2020, 35, S74–S81. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Yu, H.Q.; Sun, B.Q.; Fang, Z.F.; Zhao, J.C.; Liu, X.Y.; Li, Y.M.; Sun, X.Z.; Liang, H.F.; Zhong, B.; Huang, Z.F.; et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020. [Google Scholar] [CrossRef]
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Jiang, D.; Zhou, P.; Cheng, L.; Li, Y.; Ma, X.; Jin, T. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol. Immunol. 2020, 17, 773–775. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L.; Verdugo-Rodriguez, A.; Rodriguez, L.L.; Borca, M.V. The Role of Interleukin 6 During Viral Infections. Front. Microbiol. 2019, 10, 1057. [Google Scholar] [CrossRef] [Green Version]
- Magro, G. SARS-CoV-2 and COVID-19: Is interleukin-6 (IL-6) the ‘culprit lesion’ of ARDS onset? What is there besides Tocilizumab? SGP130Fc. Cytokine X 2020, 100029. [Google Scholar] [CrossRef]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 2011, 1813, 878–888. [Google Scholar] [CrossRef] [Green Version]
- Jurado, A.; Martin, M.C.; Abad-Molina, C.; Orduna, A.; Martinez, A.; Ocana, E.; Yarce, O.; Navas, A.M.; Trujillo, A.; Fernandez, L.; et al. COVID-19: Age, Interleukin-6, C-reactive protein, and lymphocytes as key clues from a multicentre retrospective study. Immun. Aging 2020, 17, 22. [Google Scholar] [CrossRef]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Ulhaq, Z.S.; Soraya, G.V. Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Mal. Infect. 2020, 50, 382–383. [Google Scholar] [CrossRef]
- CDC. In Vitro Diagnostics EUAs; CDC: Atlanta, GA, USA, 2020. [Google Scholar]
- Zhang, L.; Guo, H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv. Biomark. Sci. Technol. 2020, 2, 1–23. [Google Scholar] [CrossRef]
- Banerjee, N.; Mukhopadhyay, S. Viral glycoproteins: Biological role and application in diagnosis. Virusdisease 2016, 27, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.K.; Cheng, C.M. Glycan-based diagnostic devices: Current progress, challenges and perspectives. Chem. Commun. (Camb.) 2015, 51, 16750–16762. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 2020, 25. [Google Scholar] [CrossRef] [Green Version]
- Division of Viral Diseases. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2020. [Google Scholar]
- Wolfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Muller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Parikh, R.; Mathai, A.; Parikh, S.; Sekhar, G.C.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef]
- Kumar, R.; Nagpal, S.; Kaushik, S.; Mendiratta, S. COVID-19 diagnostic approaches: Different roads to the same destination. VirusDisease 2020, 31, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Wacker, M.J.; Godard, M.P. Analysis of One-Step and Two-Step Real-Time RT-PCR Using SuperScript III. J. Biomol. Tech. 2005, 16, 266–271. [Google Scholar]
- CDC. Multiplex Assay for Flu and Covid-19 & Supplies. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html (accessed on 4 November 2020).
- Van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef]
- Sethuraman, N.; Jeremiah, S.S.; Ryo, A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA 2020, 323, 2249. [Google Scholar] [CrossRef] [PubMed]
- Keikha, M. LAMP Method as One of the Best Candidates for Replacing with PCR Method. Malays. J. Med. Sci. 2018, 25, 121–123. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acid. Res. 2000, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashir, J.; Yaqinuddin, A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Klein, S.; et al. Screening for SARS-CoV-2 infections with colorimetric RT-LAMP and LAMP sequencing. medRxiv 2020. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid Detection of Novel Coronavirus (COVID-19) by Reverse Transcription-Loop- Mediated Isothermal Amplification. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Jiang, M.; Pan, W.; Arastehfar, A.; Fang, W.; ling, L.; Fang, H.; Farnaz Daneshnia, F.; Yu, J.; Liao, W.; Pei, H.; et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef] [Green Version]
- DaoThi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- Rodel, J.; Egerer, R.; Suleyman, A.; Sommer-Schmid, B.; Baier, M.; Henke, A.; Edel, B.; Loffler, B. Use of the variplex SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J. Clin. Virol. 2020, 132, 104616. [Google Scholar] [CrossRef]
- Monis, P.T.; Giglio, S. Nucleic acid amplification-based techniques for pathogen detection and identification. Infect. Genet. Evol. 2006, 6, 2–12. [Google Scholar] [CrossRef]
- Guatelli, J.C.; Whitfield, K.M.; Kwoh, D.Y.; Barringer, K.J.; Richman, D.D.; Gingeraso, T.R. Isothermal, in vitro amplification of nucleic acids by a multienzymereaction modeled after retroviral replication. Proc. Natl. Acad. Sci. USA 1990, 87, 1874–1878. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.-H. Chapter 9—Amplification of Nucleic Acids; ScienceDirect: Cambridge, MA, USA, 2019. [Google Scholar] [CrossRef]
- Gorzalski, A.J.; Tian, H.; Laverdure, C.; Morzunov, S.; Verma, S.C.; VanHooser, S.; Pandori, M.W. High-Throughput Transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020, 129, 104501. [Google Scholar] [CrossRef]
- Sarrazin, C.; Hendricks, D.A.; Sedarati, F.; Zeuzem, S. Assessment, by transcription-mediated amplification, of virologic response in patients with chronic hepatitis C virus treated with peginterferon alpha-2a. J. Clin. Microbiol. 2001, 39, 2850–2855. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.; Sahoo, M.K.; Taylor, N.; Uy, E.; Shi, R.Z.; Pinsky, B.A. Comparison of Transcription-Mediated Amplification and Real-Time PCR Assays for Hepatitis B Virus DNA Quantitation in Serum. J. Appl. Lab. Med. 2019, 4, 383–390. [Google Scholar] [CrossRef]
- Wu, S.S.; Li, Q.C.; Yin, C.Q.; Xue, W.; Song, C.Q. Advances in CRISPR/Cas-based Gene Therapy in Human Genetic Diseases. Theranostics 2020, 10, 4374–4382. [Google Scholar] [CrossRef]
- Strich, J.R.; Chertow, D.S. CRISPR-Cas Biology and Its Application to Infectious Diseases. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [Green Version]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Morisaka, H.; Yoshimi, K.; Okuzaki, Y.; Gee, P.; Kunihiro, Y.; Sonpho, E.; Xu, H.; Sasakawa, N.; Naito, Y.; Nakada, S.; et al. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat. Commun. 2019, 10, 5302. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronowski, A.M. Who or what is SHERLOCK? eJIFCC 2018, 29, 201–204. [Google Scholar]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Boehm, C.K.; Petros, B.A.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.F.; Kemball, M.E.; et al. Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yoshimi, K.; Takeshita, K.; Yamayoshi, S.; Shibumura, S.; Yamauchi, Y.; Yamamoto, M.; Yotsuyanagi, H.; Kawaoka, Y.; Mashimo, T. Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3. medRxiv 2020. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Jia, F.; Li, X.; Zhang, C.; Tang, X. The expanded development and application of CRISPR system for sensitive nucleotide detection. Protein Cell 2020. [Google Scholar] [CrossRef]
- Mota, D.S.; Marques, J.M.; Guimaraes, J.M.; Mariuba, L.A.M. Research Article CRISPR/Cas Class 2 Systems and their applications in biotechnological processes. Genet. Mol. Res. 2020, 19. [Google Scholar] [CrossRef]
- Churko, J.M.; Mantalas, G.L.; Snyder, M.P.; Wu, J.C. Overview of high throughput sequencing technologies to elucidate molecular pathways in cardiovascular diseases. Circ. Res. 2013, 112, 1613–1623. [Google Scholar] [CrossRef] [Green Version]
- Behjati, S.; Tarpey, P.S. What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 236–238. [Google Scholar] [CrossRef]
- Illumina. Comprehensive Workflow for Detecting Coronavirus Using Illumina Benchtop Systems; Illumina: San Diego, CA, USA, 2020. [Google Scholar]
- Sheridan, C. First NGS-based COVID-19 diagnostic. Nat. Biotechnol. 2020, 38, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.C.; Penrice-Randal, R.; Alruwaili, M.; Dong, X.; Pullan, S.T.; Carter, D.; Bewley, K.; Zhao, Q.; Sun, Y.; Hartley, C.; et al. Amplicon based MinION sequencing of SARS-CoV-2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID-19. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Robert, H. Enzyme-Linked Immunosorbent Assay (ELISA): A Practical Tool for Rapid Diagnosis of Viruses and Other Infectious Agents. YCBM 1979, 53, 85–92. [Google Scholar]
- Alhajj, M.; Farhana, A. Enzyme Linked Immunosorbent Assay (ELISA). Available online: https://www.ncbi.nlm.nih.gov/books/NBK555922/ (accessed on 4 November 2020).
- Gibbs, J.; Vessels, M.; Rothenberg, M. Selecting the Detection System—Colorimetric, Fluorescent, Luminescent Methods for ELISA Assays. Available online: https://www.corning.com/catalog/cls/documents/application-notes/CLS-DD-AN-458.pdf (accessed on 4 November 2020).
- Liu, T.; Zhang, J.; Yang, Y.; Ma, H.; Li, Z.; Zhang, J.; Cheng, J.; Zhang, X.; Zhao, Y.; Xia, Z.; et al. The role of interleukin-6 in monitoring severe case of coronavirus disease 2019. EMBO Mol. Med. 2020, 12, e12421. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Yan, M.; Li, H.; Liu, T.; Lin, C.; Huang, S.; Shen, C. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold- Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Du, R.H.; Li, B.; Zheng, X.S.; Yang, X.L.; Hu, B.; Wang, Y.Y.; Xiao, G.F.; Yan, B.; Shi, Z.L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [Green Version]
- Ciaurriz, P.; Fernandez, F.; Tellechea, E.; Moran, J.F.; Asensio, A.C. Comparison of four functionalization methods of gold nanoparticles for enhancing the enzyme-linked immunosorbent assay (ELISA). Beilstein J. Nanotechnol. 2017, 8, 244–253. [Google Scholar] [CrossRef] [Green Version]
- Fung, J.; Lau, S.K.P.; Woo, P.C.Y. Antigen Capture Enzyme-Linked Immunosorbent Assay for Detecting Middle East Respiratory Syndrome Coronavirus in Humans. In MERS Coronavirus: Methods and Protocols; Vijay, R., Ed.; Springer: New York, NY, USA, 2020; pp. 89–97. [Google Scholar] [CrossRef] [Green Version]
- Niikura, M.; Ikegami, T.; Saijo, M.; Kurane, I.; Miranda, M.E.; Morikawa, S. Detection of Ebola viral antigen by enzyme-linked immunosorbent assay using a novel monoclonal antibody to nucleoprotein. J. Clin. Microbiol. 2001, 39, 3267–3271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kou, G.; Dong, Y.; Zheng, Y.; Ding, Y.; Ni, W.; Wu, W.; Tang, S.; Xiong, Z.; Zhang, Y.; et al. Clinical application of Chemiluminescence Microparticle Immunoassay for SARS-CoV-2 infection diagnosis. J. Clin. Virol. 2020, 130, 104576. [Google Scholar] [CrossRef]
- Cai, X.F.; Chen, J.; Li Hu, J.; Long, Q.X.; Deng, H.J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.Z.; Wu, G.C.; et al. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 189–193. [Google Scholar] [CrossRef]
- Wu, J.L.; Tseng, W.P.; Lin, C.H.; Lee, T.F.; Chung, M.Y.; Huang, C.H.; Chen, S.Y.; Hsueh, P.R.; Chen, S.C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020. [Google Scholar] [CrossRef]
- Jiang, N.; Ahmed, R.; Damayantharan, M.; Unal, B.; Butt, H.; Yetisen, A.K. Lateral and Vertical Flow Assays for Point-of-Care Diagnostics. Adv. Healthc. Mater. 2019, 8, e1900244. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Gates-Hollingsworth, M.; Pandit, S.; Park, A.; Montgomery, D.; AuCoin, D.; Gu, J.; Zenhausern, F. Paper-based Vertical Flow Immunoassay (VFI) for detection of bio-threat pathogens. Talanta 2019, 191, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Kawde, A.-N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef] [Green Version]
- Zandian, A.; Forsstrom, B.; Haggmark-Manberg, A.; Schwenk, J.M.; Uhlen, M.; Nilsson, P.; Ayoglu, B. Whole-Proteome Peptide Microarrays for Profiling Autoantibody Repertoires within Multiple Sclerosis and Narcolepsy. J. Proteome Res. 2017, 16, 1300–1314. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.-W.; Li, Y.; Zhang, H.-N.; Wang, W.; Men, D.; Yang, X.; Qi, H.; Zhou, J.; Tao, S.-C. Global profiling of SARS-CoV-2 specific IgG/ IgM responses of convalescents using a proteome microarray. medRxiv 2020. [Google Scholar] [CrossRef]
- Dawood, A.A. Mutated COVID-19 may foretell a great risk for mankind in the future. New Microbes New Infect. 2020, 35, 100673. [Google Scholar] [CrossRef]
- Shi, H.; Han, X.; Zheng, C. Evolution of CT Manifestations in a Patient Recovered from 2019 Novel Coronavirus (2019-nCoV) Pneumonia in Wuhan, China. Radiology 2020, 295, 20. [Google Scholar] [CrossRef]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [Green Version]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Lin, B.; Zhu, Z.; Li, K.; et al. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. RSNA Radiol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, M.; Shen, C.; Wang, F.; Yuan, J.; Li, J.; Zhang, M.; Wang, Z.; Xing, L.; Wei, J.; et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Farshidfar, N.; Hamedani, S. The Potential Role of Smartphone-Based Microfluidic Systems for Rapid Detection of COVID-19 Using Saliva Specimen. Mol. Diagn. Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Caruana, G.; Croxatto, A.; Coste, A.T.; Opota, O.; Lamoth, F.; Jaton, K.; Greub, G. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol. Infect. 2020, 26, 1178–1182. [Google Scholar] [CrossRef]
- La Marca, A.; Capuzzo, M.; Paglia, T.; Roli, L.; Trenti, T.; Nelson, S.M. Testing for SARS-CoV-2 (COVID-19): A systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod. Biomed. Online 2020, 41, 483–499. [Google Scholar] [CrossRef]
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512–e00520. [Google Scholar] [CrossRef] [Green Version]
- Surveillance Guidelines for Measles, Rubella and Congenital Rubella Syndrome in the WHO European Region. Annex 3, Collection, Storage and Shipment of Specimens for Laboratory Diagnosis and Interpretation of Results. Available online: https://www.ncbi.nlm.nih.gov/books/NBK143256/ (accessed on 4 November 2020).
- Shenouda, S.Y.K. Theories of Protein Denaturation during Frozen Storage of Fish Flesh. In Advances in Food Research Volume 26; Academic Press: Cambridge, MA, USA, 1980; pp. 275–311. [Google Scholar] [CrossRef]
- Bevins, S.; Pappert, R.; Young, J.; Schmit, B.; Kohler, D.; Baeten, L. Effect of Storage Time and Storage Conditions on Antibody Detection in Blood Samples Collected on Filter Paper. J. Wildl. Dis. 2016, 52, 478–483. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]




| Techniques | Testing Materials | Platform | The Amount of Sample/Sample Size | Detection Workflow | Detection Target | Limit of Detection (LOD) |
|---|---|---|---|---|---|---|
| RT-PCR [18] | Sputum, nose, and throat swabs with and without viral transport medium | Nucleic acid | RdRp genes | 3.6 copies per reaction | ||
| E genes | 3.9 copies per reaction | |||||
| N genes | 8.3 copies per reaction | |||||
| RdRp-P2 | 21.3 copies per reaction | |||||
| RdRp-Hel | 11.2 copies per reaction | |||||
| Real time RT-PCR [19] | Tear and conjunctival secretion | Nucleic acid | 21 patients with common-type and nine patients in severe symptoms | Viral RNA in conjunctivitis | N/A | |
| RT-LAMP [20] | Viral RNA | Nucleic acid | N/A | N/A | Primer probe | LOD in the optimization: 100 copies/reaction in triplicate |
| Lateral flow immunoassay [21] | Human blood | IgG/IgM Antibody | 525 cases including 397 positive tests confirmed with PCR and 128 non-infected tests | Around 15 min | Spike protein | N/A |
| Isothermal LAMP-based [22] | Respiratory samples | Nucleic acid | ORF1ab gene | 17 copies/µL | ||
| ELISA [23] | Plasma samples from healthy blood donors, organ donors, plasma samples from upper respiratory tract swab | IgG/IgM antibody | 90 samples including 50 negative and 40 positive | 90 min for observance 5–6 h | Spike protein | |
| LFIA [23] | IgG/IgM antibody | 39–165 individual plasma samples | ||||
| Sandwich ELISA [24] | Pulmonary sarcoidosis after ablation | IgG/IgM antibody | Nucleocapsid protein | 100 ng/mL | ||
| CRISPR-Cas12 [25] | Saliva samples | Nucleic acid | RNA fragments with RdRp, ORF1b, and ORF1ab genes | ORF1ab: 10 copies/µL | ||
| Proteome Microarray [26] | Serum samples | Antibody | Antibodies | 94 pg/mL |
| Technique | Platform | Target | Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|---|---|
| PPV | NPV | |||||
| Alinity I SARS-CoV-2 IgG (Abbott) | High-throughput CMIA | Nucleocapsid | 100% | 99.9% | 84% | 100% |
| Architect SARS-CoV-2 IgG (Abbott) | High-throughput CMIA | Nucleocapsid | 100% | 99.6% | 92.9% | 100% |
| Anti-SARS-CoV-2 Rapid Test (Autobio) | Lateral flow | Spike | IgG: 99.0% IgM: 95.7% Combined: 99.0% | IgG: 99.4% IgM: 99.7% Combined: 99% | 84.4% | 99.9% |
| Platelia SARS-CoV-2 Total Ab (Bio-Rad Laboratories, Inc.) | ELISA | Nucleocapsid | 92.2% | 99.6% | 91.7% | 99.6% |
| qSARS-CoV-2 IgG/IgM Rapid Test (Cellex, Inc.) | Lateral Flow | Spike and Nucleocapsid | 93.8% | 96.0% | 55.2% | 99.7% |
| DPP COVID-19 IgM/IgG System (Chembio Diagnostic Systems, Inc.) | Lateral Flow with Reader | Nucleocapsid | IgM: 77.4% IgG: 87.1% Combined: 93.5% | Combined: 94.4% | 46.8% | 99.6% |
| LIAISON SARS-CoV-2 S1/S2 IgG (DiaSorin) | High-throughput CMIA | Spike | 97.6% | 99.3% | 88% | 99.9% |
| SARS-CoV-2 ELISA IgG (EUROIMMUN) | ELISA | Spike | 90.0% | 100% | 100% | 99.5% |
| COVID-19 ELISA Antibody Test (Mount Sinai Hospital Clinical Laboratory) | 2-Step ELISA | Spike | Combined: 92.5% | 100% | 100% | 99.6% |
| VITROS Anti-SARS-CoV-2 IgG test (Ortho-Clinical Diagnostics, Inc.) | High-Throughput CLIA | Spike | IgG: 90.0% | IgG: 100% | 100% | 99.5% |
| VITROS Immunodiagnostic Products Anti-SARS-CoV-2 Total Reagent Pack and Calibrator (Ortho-Clinical Diagnostics, Inc.) | High-Throughput CLIA | Spike | 100% | 100% | 100% | 100% |
| Elecsys Anti-SARS-CoV-2 (Roche) | High-Throughput ECLIA | Nucleocapsid | 100% | 99.8% | 96.5% | 100% |
| New York SARS-CoV Microsphere Immunoassay for Antibody Detection (Wadsworth Center, New York State Department of Health) | MIA | Nucleocapsid | 88% | 98.8% | 79.4 | 99.4% |
| Organization | Test Name | Test Result Time | Specimen Type | LOD | Characteristics |
|---|---|---|---|---|---|
| Access Bio, Inc. | CareStart COVID-19 Antigen test | 10 min | Direct Nasopharyngeal Swab | 8 × 102 TCID50/mL | SARS-CoV-2 Nucleocapsid Protein-Specific, Lateral Flow, Visual Readout, Lab-based, Point-of-Care Testing (POC) but requires a sample preparation step and a trained operator to perform the test. |
| Quidel Corporation | Sofia 2 Flu + SARS Antigen FIA | 15 min | Direct Nasal or Nasopharyngeal Swabs | 4.17 × 105 TCID50/mL | Mulitiplex Detection (SARS-CoV-2, Influenza A Virus, and Influenza B Virus), Nucleocapsid Protein-Specific, SARS-CoV, and SARS-CoV-2 Detection but cannot differentiate between them, Lateral Flow, Instrument-based Immunofluorescence Read, Lab-based, POC Testing but is limited to Sofia 2 Instrument for results, Trained Operator required. |
| Abbott Diagnostics Scarborough, Inc. | BinaxNOW COVID-19 Ag Card | 15 min | Direct Nasal Swab | 22.5 TCID50/swab | Singleplex SARS-CoV-2 Detection, No Specific to SARS-CoV-2 Detection alone (SARS-CoV Cross-reaction), Nucleocapsid Protein-Specific, Lateral Flow Immunoassay, Visual Colorimetric Pink/Purple Read but results interpretation is limited to individuals with color-impaired vision, POCT Testing but testing performance is limited by following meticulous testing instructions. |
| Lumira Dx UK Ltd. | Lumira Dx SARS-CoV-2 Ag Test | 12 min | Direct Nasal Swab | 32 TCID50/mL | Single-Use Microfluidic Fluorescence Immunoassay, SARS-CoV-2 Nucleocapsid Protein-Specific, Digital Instrument Read, Small Sample Testing Volume (one drop), POC Testing but limited to Healthcare Professional-Use-Only proficient in performing tests using the Lumira Dx Platform, Requires Lumira Dx Test Strip. |
| Becton, Dickinson, and Company (BD) | BD Veritor System for Rapid SARS-CoV-2 Detection | 15 min | Direct Nasal Swab | 1.4 × 102 TCID50/mL | Chromatographic Immunoassay, SARS-CoV-2 Nucleocapsid Antigen-Specific, Digital Instrument Reading, Result Documentation Capabilities, Good Analytical Sensitivity, Laboratory-based, POC Testing but is limited to patient testing environments where test interpretation can only be done with the BD Veritor Plus Analyzer Instrument. |
| Quidel Corporation | Sofia SARS Antigen FIA | 16 min | Direct Nasal or Nasopharyngeal Swabs | 1.13 × 102 TCID50/mL | Immunofluorescent Sandwich Assay, Lateral Flow, Singleplex Detection, SARS-CoV or SARS-CoV-2 Nucleocapsid Antigen-Specific but does not differentiate between them, Instrument Reading, Good Analytical Sensitivity, Results Affected by High Viscous Samples, Result interpretation limited to trained clinical laboratory personnel proficient in performing tests using Sofia and Sofia 2 instruments. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.N.T.; McCarthy, C.; Lantigua, D.; Camci-Unal, G. Development of Diagnostic Tests for Detection of SARS-CoV-2. Diagnostics 2020, 10, 905. https://doi.org/10.3390/diagnostics10110905
Nguyen NNT, McCarthy C, Lantigua D, Camci-Unal G. Development of Diagnostic Tests for Detection of SARS-CoV-2. Diagnostics. 2020; 10(11):905. https://doi.org/10.3390/diagnostics10110905
Chicago/Turabian StyleNguyen, Ngan N. T., Colleen McCarthy, Darlin Lantigua, and Gulden Camci-Unal. 2020. "Development of Diagnostic Tests for Detection of SARS-CoV-2" Diagnostics 10, no. 11: 905. https://doi.org/10.3390/diagnostics10110905
APA StyleNguyen, N. N. T., McCarthy, C., Lantigua, D., & Camci-Unal, G. (2020). Development of Diagnostic Tests for Detection of SARS-CoV-2. Diagnostics, 10(11), 905. https://doi.org/10.3390/diagnostics10110905





