A ‘Real-Life’ Experience on Automated Digital Image Analysis of FGFR2 Immunohistochemistry in Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Samples and Immunohistochemistry for FGFR2
2.2. Digitalization, Manual and Automated Assessment of FGFR2 Staining
2.3. Statistical Analysis of Reliability between Automated and Expert Evaluation
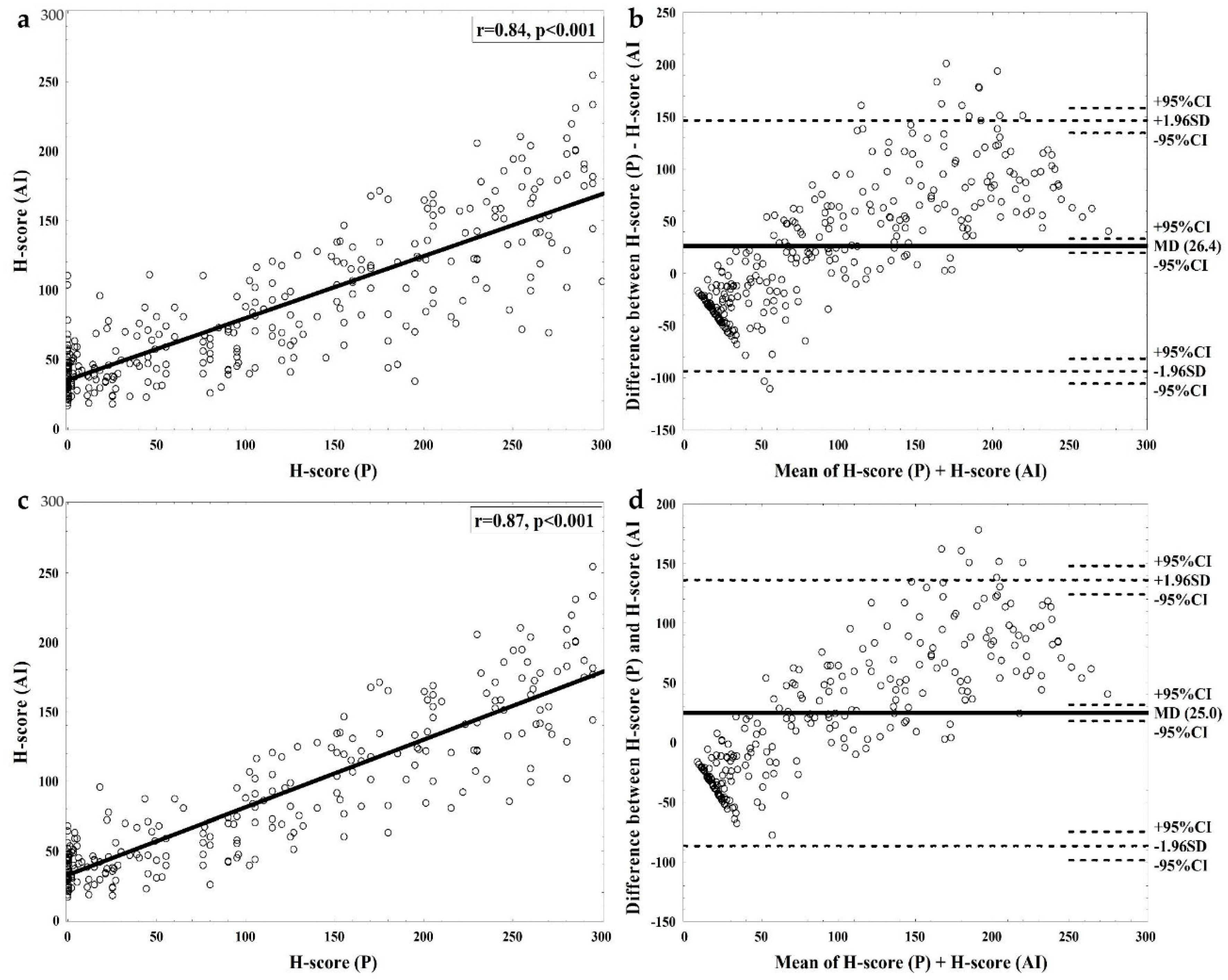
3. Results
3.1. Interrater Agreement between Pathologist’s and Software Assessment
3.2. Sources of Discrepancy between Pathologist’s and Software Assessment
3.3. Time of Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Grzybowski, A.; Brona, P.; Lim, G.; Ruamviboonsuk, P.; Tan, G.S.W.; Abramoff, M.; Ting, D.S.W. Artificial intelligence for diabetic retinopathy screening: A review. Eye 2020, 34, 451–460. [Google Scholar] [CrossRef]
- Yu, K.-H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology-new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and Deep Learning in Diagnostic Pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef]
- Bulten, W.; Pinckaers, H.; Van Boven, H.; Vink, R.; De Bel, T.; Van Ginneken, B.; Van Der Laak, J.; Hulsbergen-Van De Kaa, C.; Litjens, G. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study. Lancet Oncol. 2020, 21, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes Van Diest, P.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199. [Google Scholar] [CrossRef]
- Vandenberghe, M.E.; Scott, M.L.J.; Scorer, P.W.; Söderberg, M.; Balcerzak, D.; Barker, C. Relevance of deep learning to facilitate the diagnosis of HER2 status in breast cancer. Sci. Rep. 2017, 7, 45938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushnarev, V.A.; Artemyeva, E.S.; Kudaybergenova, A.G. Comparison of digital and visual methods for Ki-67 assessment in invasive breast carcinomas. Arkhiv Patol. 2018, 80, 38. [Google Scholar] [CrossRef] [PubMed]
- Acs, B.; Pelekanou, V.; Bai, Y.; Martinez-Morilla, S.; Toki, M.; Leung, S.C.Y.; Nielsen, T.O.; Rimm, D.L. Ki67 reproducibility using digital image analysis: An inter-platform and inter-operator study. Lab. Investig. 2019, 99, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, D.; Braun, M.; Kitowska, K.; Mieczkowski, K.; Kordek, R.; Sadej, R.; Romanska, H. FGFs/FGFRs-dependent signalling in regulation of steroid hormone receptors–implications for therapy of luminal breast cancer. J. Exp. Clin. Cancer Res. 2019, 38, 230. [Google Scholar] [CrossRef] [Green Version]
- Sengal, A.T.; Patch, A.M.; Snell, C.E.; Smith, D.S.; Leung, S.C.Y.; Talhouk, A.; Williams, E.D.; McAlpine, J.N.; Pollock, P.M. FGFR2c Mesenchymal Isoform Expression Is Associated with Poor Prognosis and Further Refines Risk Stratification within Endometrial Cancer Molecular Subtypes. Clin. Cancer Res. 2020, 26, 4569–4580. [Google Scholar] [CrossRef]
- Martin, A.J.; Grant, A.; Ashfield, A.M.; Palmer, C.N.; Baker, L.; Quinlan, P.R.; Purdie, C.A.; Thompson, A.M.; Jordan, L.B.; Berg, J.N. FGFR2 protein expression in breast cancer: Nuclear localisation and correlation with patient genotype. BMC Res. Notes 2011, 4, 72. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.; Lee, J.; Hong, M.; Kim, S.T.; Park, S.H.; Choi, M.G.; Lee, J.H.; Sohn, T.S.; Bae, J.M.; Kim, S.; et al. FGFR2 in gastric cancer: Protein overexpression predicts gene amplification and high H-index predicts poor survival. Mod. Pathol. 2016, 29, 1095–1103. [Google Scholar] [CrossRef]
- Kurban, G.; Ishiwata, T.; Kudo, M.; Yokoyama, M.; Sugisaki, Y.; Naito, Z. Expression of keratinocyte growth factor receptor (KGFR/FGFR2 IIIb) in human uterine cervical cancer. Oncol. Rep. 2004, 11, 987–991. [Google Scholar] [CrossRef]
- Braun, M.; Piasecka, D.; Tomasik, B.; Mieczkowski, K.; Stawiski, K.; Zielinska, A.; Kopczynski, J.; Nejc, D.; Kordek, R.; Sadej, R.; et al. Hormonal Receptor Status Determines Prognostic Significance of FGFR2 in Invasive Breast Carcinoma. Cancers 2020, 12, 2713. [Google Scholar] [CrossRef]
- Babina, I.S.; Turner, N.C. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 2017, 17, 318–332. [Google Scholar] [CrossRef]
- Hunter, D.J.; Kraft, P.; Jacobs, K.B.; Cox, D.G.; Yeager, M.; Hankinson, S.E.; Wacholder, S.; Wang, Z.; Welch, R.; Hutchinson, A.; et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007, 39, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, J.F.; Kornegoor, R.; Van Der Wall, E.; Van Der Groep, P.; Van Diest, P.J. Differential Expression of Growth Factor Receptors and Membrane-Bound Tumor Markers for Imaging in Male and Female Breast Cancer. PLoS ONE 2013, 8, e53353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piasecka, D.; Kitowska, K.; Czaplinska, D.; Mieczkowski, K.; Mieszkowska, M.; Turczyk, L.; Skladanowski, A.C.; Zaczek, A.J.; Biernat, W.; Kordek, R.; et al. Fibroblast growth factor signalling induces loss of progesterone receptor in breast cancer cells. Oncotarget 2016, 7, 86011–86025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czaplinska, D.; Mieczkowski, K.; Supernat, A.; Skladanowski, A.C.; Kordek, R.; Biernat, W.; Zaczek, A.J.; Romanska, H.M.; Sadej, R. Interactions between FGFR2 and RSK2—Implications for breast cancer prognosis. Tumour Biol. 2016, 37, 13721–13731. [Google Scholar] [CrossRef] [Green Version]
- Campbell, T.M.; Castro, M.A.A.; de Santiago, I.; Fletcher, M.N.C.; Halim, S.; Prathalingam, R.; Ponder, B.A.J.; Meyer, K.B. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis 2016, 37, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Wu, D.; Wang, W.; He, X.; Wang, M. Variants of FGFR2 and their associations with breast cancer risk: A HUGE systematic review and meta-analysis. Breast Cancer Res. Treat. 2016, 155, 313–335. [Google Scholar] [CrossRef]
- Turczyk, L.; Kitowska, K.; Mieszkowska, M.; Mieczkowski, K.; Czaplinska, D.; Piasecka, D.; Kordek, R.; Skladanowski, A.C.; Potemski, P.; Romanska, H.M.; et al. FGFR2-Driven Signaling Counteracts Tamoxifen Effect on ERalpha-Positive Breast Cancer Cells. Neoplasia 2017, 19, 791–804. [Google Scholar] [CrossRef]
- Campbell, T.M.; Castro, M.A.A.; De Oliveira, K.G.; Ponder, B.A.J.; Meyer, K.B. ERα Binding by Transcription Factors NFIB and YBX1 Enables FGFR2 Signaling to Modulate Estrogen Responsiveness in Breast Cancer. Cancer Res. 2018, 78, 410–421. [Google Scholar] [CrossRef] [Green Version]
- Sinn, H.P.; Kreipe, H. A Brief Overview of the WHO Classification of Breast Tumors, 4th Edition, Focusing on Issues and Updates from the 3rd Edition. Breast Care 2013, 8, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Hoon Tan, P.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 WHO classification of tumours of the breast. Histopathology 2020. [Google Scholar] [CrossRef]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilic-Zulle, L. Comparison of methods: Passing and Bablok regression. Biochem. Med. 2011, 21, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Lebeau, A.; Bu, H.; Hartmann, A.; Penault-Llorca, F.; Guerini-Rocco, E.; Schraml, P.; Symmans, F.; Stoehr, R.; Teng, X.; et al. An international reproducibility study validating quantitative determination of ERBB2, ESR1, PGR, and MKI67 mRNA in breast cancer using MammaTyper®. Breast Cancer Res. 2017, 19, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, V.S.; Ficsor, L.; Kamaras, V.; Jonas, V.; Virag, T.; Tulassay, Z.; Molnar, B. Automated multichannel fluorescent whole slide imaging and its application for cytometry. Cytom. A 2009, 75, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Rojo, M.G.; Bueno, G.; Slodkowska, J. Review of imaging solutions for integrated quantitative immunohistochemistry in the Pathology daily practice. Folia Histochem. Cytobiol. 2009, 47, 349–354. [Google Scholar] [CrossRef] [Green Version]
- Granter, S.R.; Beck, A.H.; Papke, D.J. AlphaGo, Deep Learning, and the Future of the Human Microscopist. Arch. Pathol. Lab. Med. 2017, 141, 619–621. [Google Scholar] [CrossRef] [Green Version]
- Jha, S.; Topol, E.J. Adapting to Artificial Intelligence. JAMA 2016, 316, 2353. [Google Scholar] [CrossRef]
- Waszczykowska, A.; Zmysłowska, A.; Braun, M.; Zielonka, E.; Ivask, M.; Koks, S.; Jurowski, P.; Młynarski, W. Corneal Abnormalities Are Novel Clinical Feature in Wolfram Syndrome. Am. J. Ophthalmol. 2020, 217, 140–151. [Google Scholar] [CrossRef]
- Piasecka, D.; Braun, M.; Mieszkowska, M.; Kowalczyk, L.; Kopczynski, J.; Kordek, R.; Sadej, R.; Romanska, H.M. Upregulation of HIF1-α via an NF-κB/COX2 pathway confers proliferative dominance of HER2-negative ductal carcinoma in situ cells in response to inflammatory stimuli. Neoplasia 2020, 22, 576–589. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Braun, M.; Piasecka, D.; Bobrowski, M.; Kordek, R.; Sadej, R.; Romanska, H.M. A ‘Real-Life’ Experience on Automated Digital Image Analysis of FGFR2 Immunohistochemistry in Breast Cancer. Diagnostics 2020, 10, 1060. https://doi.org/10.3390/diagnostics10121060
Braun M, Piasecka D, Bobrowski M, Kordek R, Sadej R, Romanska HM. A ‘Real-Life’ Experience on Automated Digital Image Analysis of FGFR2 Immunohistochemistry in Breast Cancer. Diagnostics. 2020; 10(12):1060. https://doi.org/10.3390/diagnostics10121060
Chicago/Turabian StyleBraun, Marcin, Dominika Piasecka, Mateusz Bobrowski, Radzislaw Kordek, Rafal Sadej, and Hanna M. Romanska. 2020. "A ‘Real-Life’ Experience on Automated Digital Image Analysis of FGFR2 Immunohistochemistry in Breast Cancer" Diagnostics 10, no. 12: 1060. https://doi.org/10.3390/diagnostics10121060




