Carotid Artery Plaque Identification and Display System (MRI-CAPIDS) Using Opensource Tools
Abstract
:1. Introduction
- The loading of the images is not dependent on the specific names of the folders for each DICOM image type as long as each of the five folders contain valid DICOM files. All the labels/information displayed for each sub-window are extracted from the DICOM files themselves.
- Minimum 10,000% factor improvement in QImage loading for all the MRI image types in the modified code compared to the routine in the existing original QDCM library.
- In terms of system development, all the sub-windows for each of the MRI image type are inherited from a parent sub-window which makes it easier for the developer to write specific functions for the MRI images.
Background
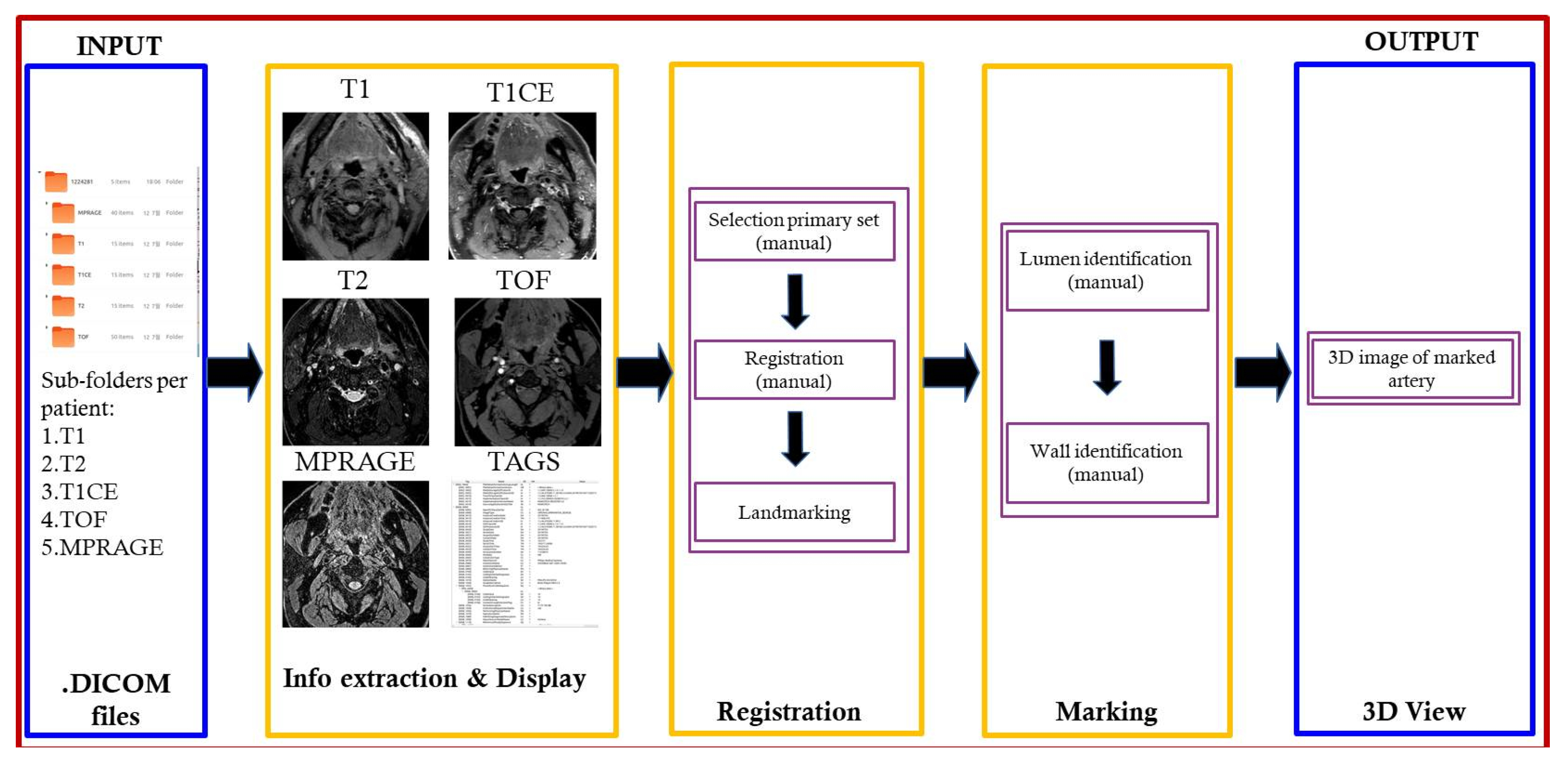
2. Proposed Architecture
2.1. Research Purpose
2.2. Algorithm
3. Open Source Tools
3.1. Qt-Anywhere on Ubuntu Linux
3.2. DICOM and QDCM Library
- A Tag (Figure 2): identifies the attribute; usually in (XXXX,XXXX) hexadecimal format; can be further split into group number and element number.
- A Name: descriptive text that describes the tag.
- A Value Representation (VR): identifies the data type and format of the attribute value.
- A Value Multiplicity (VM): defines whether an attribute can or cannot include multiple elements.
- The Value: the attribute itself.
3.3. OpenGL
4. Design and Development
- Browse, open, and load MRI files from patient folder.
- Select MRI image file to be set as the primary image set (T1, T2, T1CE, TOF, MPRAGE).
- Rationalize the image set by ensuring that all image types are slice aligned and include the same number of images per set for processing.
- Identify by marking lumen and wall for each image slice.
- Generate 3D rendering of marked lumen and wall.
4.1. Image/Data Loading Module
Faster Image Loading
4.2. Registration Module
4.3. Marking Module
4.4. 3D View Module
4.4.1. Smoother Contour of Polygon
4.4.2. Generation of the 3D View
5. Resulting System
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kerwin, W.S.; Han, C.; Chu, B.; Xu, D.; Luo, Y.; Hwang, J.-N.; Hatsukami, T.; Yuan, C. A Quantitative Vascular Analysis System for Evaluation of Atherosclerotic Lesions by MRI. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2001, Utrecht, The Netherlands, 14–17 October 2001; pp. 786–794. [Google Scholar]
- Fuster, V.; Stein, B.; Ambrose, J.A.; Badimon, L.; Badimon, J.J.; Chesebro, J.H. Atherosclerotic plaque rupture and thrombosis. Evolving concepts. Circulation 1990, 82, II47–II59. [Google Scholar] [PubMed]
- Fayad, Z.; Fuster, V. Characterization of atherosclerotic plaques by magnetic resonance imaging. Ann. N. Y. Acad. Sci. 2000, 902, 173–186. [Google Scholar] [PubMed]
- Von Ingersleben, G.; Schmiedl, U.P.; Hatsukami, T.S.; Nelson, J.A.; Subramaniam, D.S.; Ferguson, M.S.; Yuan, C. Characterization of atherosclerotic plaques at the carotid bifurcation: Correlation of high-resolution MR imaging with histologic analysis—Preliminary study. Radiographics 1997, 17, 1417–1423. [Google Scholar] [PubMed] [Green Version]
- Hatsukami, T.S.; Ross, R.; Polissar, N.L.; Yuan, C. Visualization of Fibrous Cap Thickness and Rupture in Human Atherosclerotic Carotid Plaque In Vivo With High-Resolution Magnetic Resonance Imaging. Circulation 2000, 102, 959–964. [Google Scholar]
- Yuan, C.; Beach, K.; Smith, H.; Hatsukami, T. In vivo measurements of maximum plaque area based on high resolution MRI. Circulation 1998, 98, 2666–2671. [Google Scholar]
- Henry, R.; Kostense, P.J.; Dekker, J.M.; Nijpels, G.; Heine, R.J.; Kamp, O.; Bouter, L.M.; Stehouwer, C.D. Carotid Arterial Remodeling. Stroke 2004, 35, 671–676. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.-M.; Hatsukami, T.S.; Ferguson, M.S.; Small, R.; Polissar, N.L.; Yuan, C. Classification of Human Carotid Atherosclerotic Lesions With In Vivo Multicontrast Magnetic Resonance Imaging. Circulation 2002, 106, 1368–1373. [Google Scholar]
- Stary, H.C.; Chandler, A.B.; Dinsmore, R.E.; Fuster, V.; Glagov, S.; Insull, W.; Rosenfeld, M.E.; Schwartz, C.J.; Wagner, W.D.; Wissler, R.W. A Definition of Advanced Types of Atherosclerotic Lesions and a Histological Classification of Atherosclerosis. Circulation 1995, 92, 1355–1374. [Google Scholar] [CrossRef]
- Stary, H.C. Natural History and Histological Classification of Atherosclerotic Lesions. Arter. Thromb. Vasc. Biol. 2000, 20, 1177–1178. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Oikawa, M.; Miller, Z.; Hatsukami, T. MRI of carotid atherosclerosis. J. Nucl. Cardiol. 2008, 15, 266–275. [Google Scholar]
- Kerwin, W.; Xu, D.; Liu, F.; Saam, T.; Underhill, H.; Takaya, N.; Baocheng, C.; Thomas, H.; Chun, Y. Magnetic resonance imaging of carotid atherosclerosis: Plaque analysis. Top. Magn. Reson. Imaging 2007, 18, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Commandeur, F.; Goeller, M.; Dey, D. Cardiac CT: Technological Advances in Hardware, Software, and Machine Learning Applications. Curr. Cardiovasc. Imaging Rep. 2018, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, S.; Moselewski, F.; Ropers, D.; Ferencik, M.; Hoffmann, U.; MacNeill, B.; Pohle, K.; Baum, U.; Anders, K.; Jang, I.-k.; et al. Detection of calcified and noncalcified coronary atherosclerotic plaque by contrast-enhanced, submillimeter multidetector spiral computed tomography: A segment-based comparison with intravascular ultrasound. Circulation 2004, 109, 14–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pundziute, G.; Schuijf, J.D.; Jukema, J.W.; Decramer, I.; Sarno, G.; Vanhoenacker, P.K.; Boersma, E.; Reiber, J.H.; Schalij, M.J.; Wijns, W.; et al. Evaluation of plaque characteristics in acute coronary syndromes: Non-invasive assessment with multi-slice computed tomography and invasive evaluation with intravascular ultrasound radiofrequency data analysis. Eur. Heart J. 2008, 29, 2373–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harteveld, A.A.; Van Der Kolk, A.G.; Zwanenburg, J.J.; Luijten, P.R.; Hendrikse, J. 7-T MRI in Cerebrovascular Diseases. Top. Magn. Reson. Imaging 2016, 25, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Johst, S.; Wrede, K.H.; Ladd, M.E.; Maderwald, S. Time-of-Flight Magnetic Resonance Angiography at 7 T Using Venous Saturation Pulses with Reduced Flip Angles. Investig. Radiol. 2012, 47, 445–450. [Google Scholar] [CrossRef]
- Wrede, K.H.; Dammann, P.; Mönninghoff, C.; Johst, S.; Maderwald, S.; Sandalcioglu, I.E.; Müller, O.; Özkan, N.; Ladd, M.E.; Forsting, M.; et al. Non-Enhanced MR Imaging of Cerebral Aneurysms: 7 Tesla versus 1.5 Tesla. PLoS ONE 2014, 9, e84562. [Google Scholar] [CrossRef]
- Hendrikse, J.; Zwanenburg, J.J.; Visser, F.; Takahara, T.; Luijten, P. Noninvasive Depiction of the Lenticulostriate Arteries with Time-of-Flight MR Angiography at 7.0 T. Cerebrovasc. Dis. 2008, 26, 624–629. [Google Scholar] [CrossRef]
- Zwanenburg, J.J.; Hendrikse, J.; Takahara, T.; Visser, F.; Luijten, P.R. MR angiography of the cerebral perforating arteries with magnetization prepared anatomical reference at 7T: Comparison with time-of-flight. J. Magn. Reson. Imaging 2008, 28, 1519–1526. [Google Scholar] [CrossRef]
- Koning, W.; Bluemink, J.J.; Langenhuizen, E.A.J.; Raaijmakers, A.J.; Andreychenko, A.; Berg, C.A.T.V.D.; Luijten, P.R.; Zwanenburg, J.J.; Klomp, D.W.J. High-resolution MRI of the carotid arteries using a leaky waveguide transmitter and a high-density receive array at 7 T. Magn. Reson. Med. 2012, 69, 1186–1193. [Google Scholar] [CrossRef]
- Hartog, A.D.; Bovens, S.; Koning, W.; Hendrikse, J.; Luijten, P.; Moll, F.; Pasterkamp, G.; De Borst, G. Current Status of Clinical Magnetic Resonance Imaging for Plaque Characterisation in Patients with Carotid Artery Stenosis. Eur. J. Vasc. Endovasc. Surg. 2013, 45, 7–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseini, A.A.; Kandiyil, N.; MacSweeney, S.T.; Altaf, N.; Auer, D.P. Carotid plaque hemorrhage on magnetic resonance imaging strongly predicts recurrent ischemia and stroke. Ann. Neurol. 2013, 73, 774–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Moody, A.R.; Roifman, I.; Bluemke, D.A.; Zavodni, A. Advanced MRI for carotid plaque imaging. Int. J. Cardiovasc. Imaging 2016, 32, 83–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoff, M.N.; McKinney, A.M.; Shellock, F.G.; Rassner, U.; Gilk, T.; Watson, R.E.; Greenberg, T.D.; Froelich, J.; Kanal, E. Safety Considerations of 7-T MRI in Clinical Practice. Radiology 2019, 292, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Hansson, B.; Bloch, K.M.; Owman, T.; Nilsson, M.; Lätt, J.; Olsrud, J.; Björkman-Burtscher, I.M. Subjectively Reported Effects Experienced in an Actively Shielded 7T MRI: A Large-Scale Study. J. Magn. Reson. Imaging 2020, 52, 1265–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardinal, M.-H.R.; Heusinkveld, M.H.G.; Qin, Z.; Lopata, R.G.P.; Naim, C.; Soulez, G.; Cloutier, G. Carotid Artery Plaque Vulnerability Assessment Using Noninvasive Ultrasound Elastography: Validation with MRI. Am. J. Roentgenol. 2017, 209, 142–151. [Google Scholar] [CrossRef]
- Johri, A.M.; Nambi, V.; Naqvi, T.Z.; Feinstein, S.B.; Kim, E.S.; Park, M.M.; Becher, H.; Sillesen, H. Recommendations for the Assessment of Carotid Arterial Plaque by Ultrasound for the Characterization of Atherosclerosis and Evaluation of Cardiovascular Risk: From the American Society of Echocardiography. J. Am. Soc. Echocardiogr. 2020, 33, 917–933. [Google Scholar] [CrossRef]
- Biermann, C.; Tsiflikas, I.; Thomas, C.; Kasperek, B.; Heuschmid, M.; Claussen, C.D. Evaluation of Computer-Assisted Quantification of Carotid Artery Stenosis. J. Digit. Imaging 2012, 25, 250–257. [Google Scholar] [CrossRef] [Green Version]
- Siemens-Healthineers. Syngo Circulation Plaque Analysis. Computed Tomography—Clinical Software Application. Available online: https://www.siemens-healthineers.com/computed-tomography/options-upgrades/clinical-applications/syngo-circulation-plaque-analysis/features (accessed on 3 December 2019).
- Medis Medical Imaging Systems bv Company. Plaque Burden. Plaque Burden powered by QAngio CT. Available online: https://www.medis.nl/apps/plaque-burden/ (accessed on 3 December 2019).
- ITK-SNAP Home. Available online: http://www.itksnap.org/pmwiki/pmwiki.php (accessed on 29 June 2020).
- 3D Slicer. Available online: https://www.slicer.org/ (accessed on 29 June 2020).
- Ginkgo CADx|Ginkgo CADx, Open Core DICOM Viewer + Dicomizer + CADx. Available online: http://ginkgo-cadx.com/en/ (accessed on 29 June 2020).
- Index of /archive/qt/5.12/5.12.3. Available online: https://download.qt.io/archive/qt/5.12/5.12.3/ (accessed on 29 June 2020).
- DICOM Standard. Available online: https://www.dicomstandard.org/ (accessed on 29 June 2020).
- QDCM: DICOM for Qt. Available online: http://qdcm.sourceforge.net/ (accessed on 29 June 2020).
- OpenGL—The Industry Standard for High Performance Graphics. Available online: https://www.opengl.org/ (accessed on 29 June 2020).
- Jodas, D.S.; Pereira, A.S.; Tavares, J.M.R.S. A review of computational methods applied for identification and quantification of atherosclerotic plaques in images. Expert Syst. Appl. 2016, 46, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Uc, B.M.; Castillo-Sánchez, G.; Marques, G.; Arambarri, J.; De La Torre-Díez, I. An Experience of Electronic Health Records Implementation in a Mexican Region. J. Med. Syst. 2020, 44, 106. [Google Scholar] [CrossRef]








| Conventional AHA Classification (Stary) | Modified AHA Classification for MRI (Cai et al. [8]) |
|---|---|
| Type I: initial lesion with foam cells | Type I–II: near-normal wall thickness, no calcification |
| Type II: fatty streak with multiple foam cell layers | |
| Type III: preatheroma with extracellular lipid pools | Type III: diffuse intimal thickening or small eccentric plaque with no calcification |
| Type IV: atheroma with a confluent extracellular lipid core | Type IV–V: plaque with a lipid or necrotic core surrounded by fibrous tissue with possible calcification |
| Type V: fibroatheroma | |
| Type VI: complex plaque with possible surface defect, hemorrhage, or thrombus | Type VI: complex plaque with possible surface defect, hemorrhage, or thrombus |
| Type VII: calcified plaque | Type VII: calcified plaque |
| Type VIII: fibrotic plaque without lipid core | Type VIII: fibrotic plaque without lipid core and with possible small calcifications |
| Image to QImage Conversion Algorithm | MRI Types | ||||
|---|---|---|---|---|---|
| 3D MP-RAGE_UW_d8000 | T1 FS TSE BB | T1 FS TSE BB CM | T2 FS TSE BB | 3D TOF Neck | |
| Original QDCM | 74.00 ms | 249.67 ms | 249 ms | 244.33 ms | 280.33 ms |
| Modified | 0.67 ms | 2.33 ms | 1.67 ms | 2.00 ms | 1.67 ms |
| Factor Improvement | 11,100.00% | 10,700.00% | 14,940.00% | 12,216.67% | 16,820.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vista, F.P., IV; Ngo, M.T.; Cho, S.B.; Kwak, H.S.; Chong, K.T. Carotid Artery Plaque Identification and Display System (MRI-CAPIDS) Using Opensource Tools. Diagnostics 2020, 10, 1111. https://doi.org/10.3390/diagnostics10121111
Vista FP IV, Ngo MT, Cho SB, Kwak HS, Chong KT. Carotid Artery Plaque Identification and Display System (MRI-CAPIDS) Using Opensource Tools. Diagnostics. 2020; 10(12):1111. https://doi.org/10.3390/diagnostics10121111
Chicago/Turabian StyleVista, Felipe P., IV, Minh Tri Ngo, Seung Bin Cho, Hyo Sung Kwak, and Kil To Chong. 2020. "Carotid Artery Plaque Identification and Display System (MRI-CAPIDS) Using Opensource Tools" Diagnostics 10, no. 12: 1111. https://doi.org/10.3390/diagnostics10121111
APA StyleVista, F. P., IV, Ngo, M. T., Cho, S. B., Kwak, H. S., & Chong, K. T. (2020). Carotid Artery Plaque Identification and Display System (MRI-CAPIDS) Using Opensource Tools. Diagnostics, 10(12), 1111. https://doi.org/10.3390/diagnostics10121111








