Development of a Deep-Learning-Based Artificial Intelligence Tool for Differential Diagnosis between Dry and Neovascular Age-Related Macular Degeneration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Subjects
2.3. Imaging Equipment
2.4. Convolutional Neural Network (CNN) Modeling
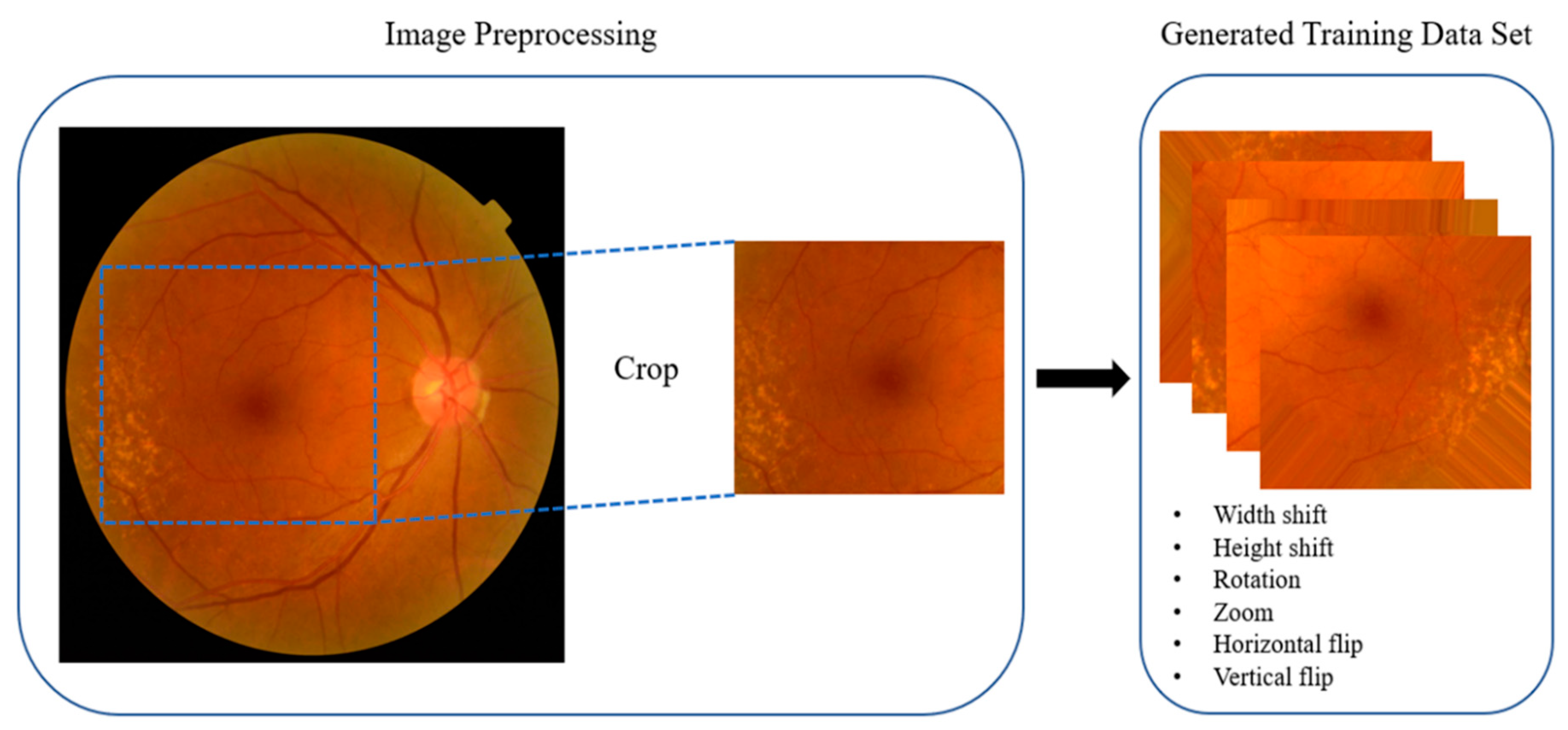
2.5. Preprocessing
2.6. Cross-Validation of Artificial Intelligence (AI)-Based Diagnosis
2.7. Comparative Analysis of Accuracy Values of the AI Diagnosis Tool and Residents in Ophthalmology
3. Results
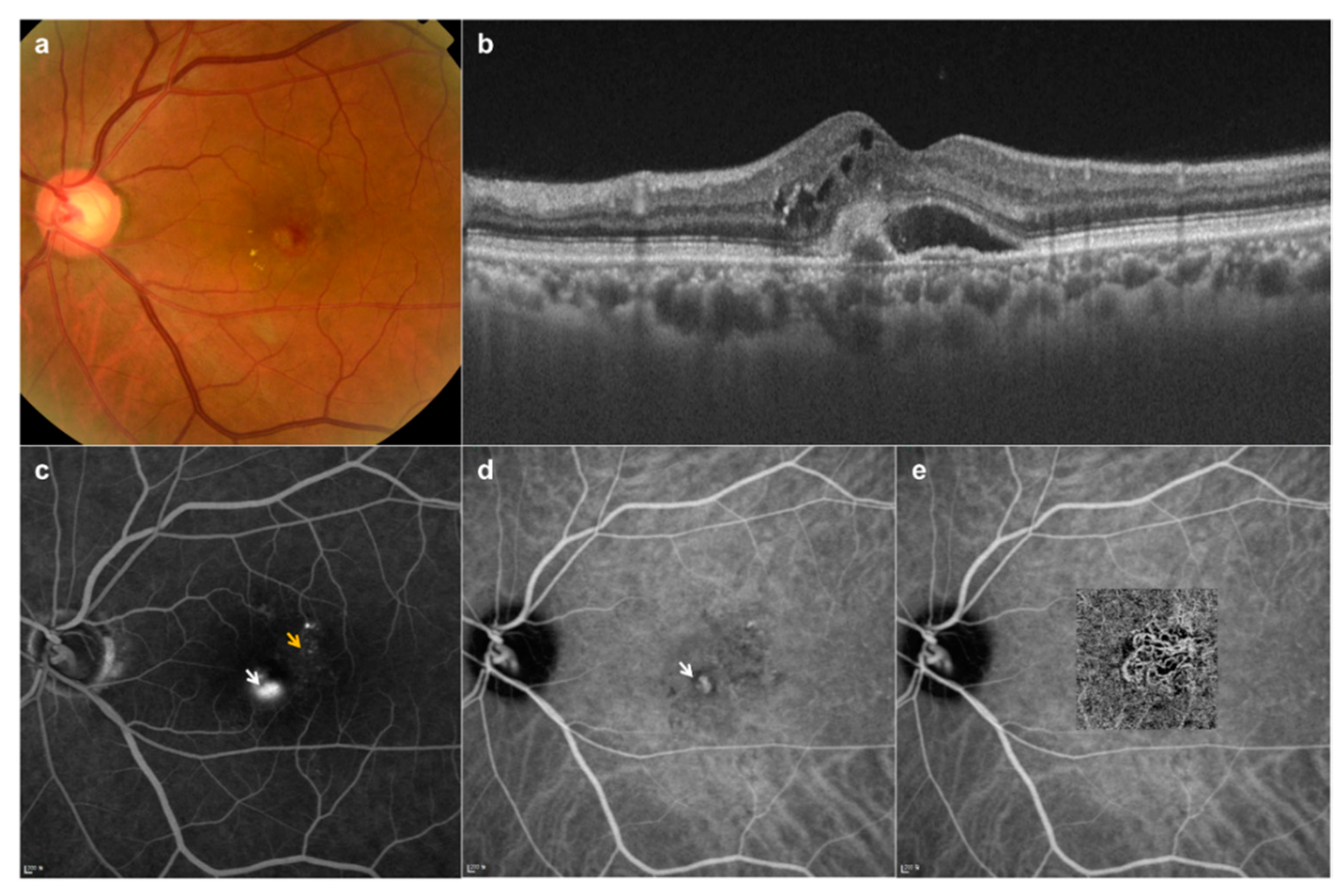
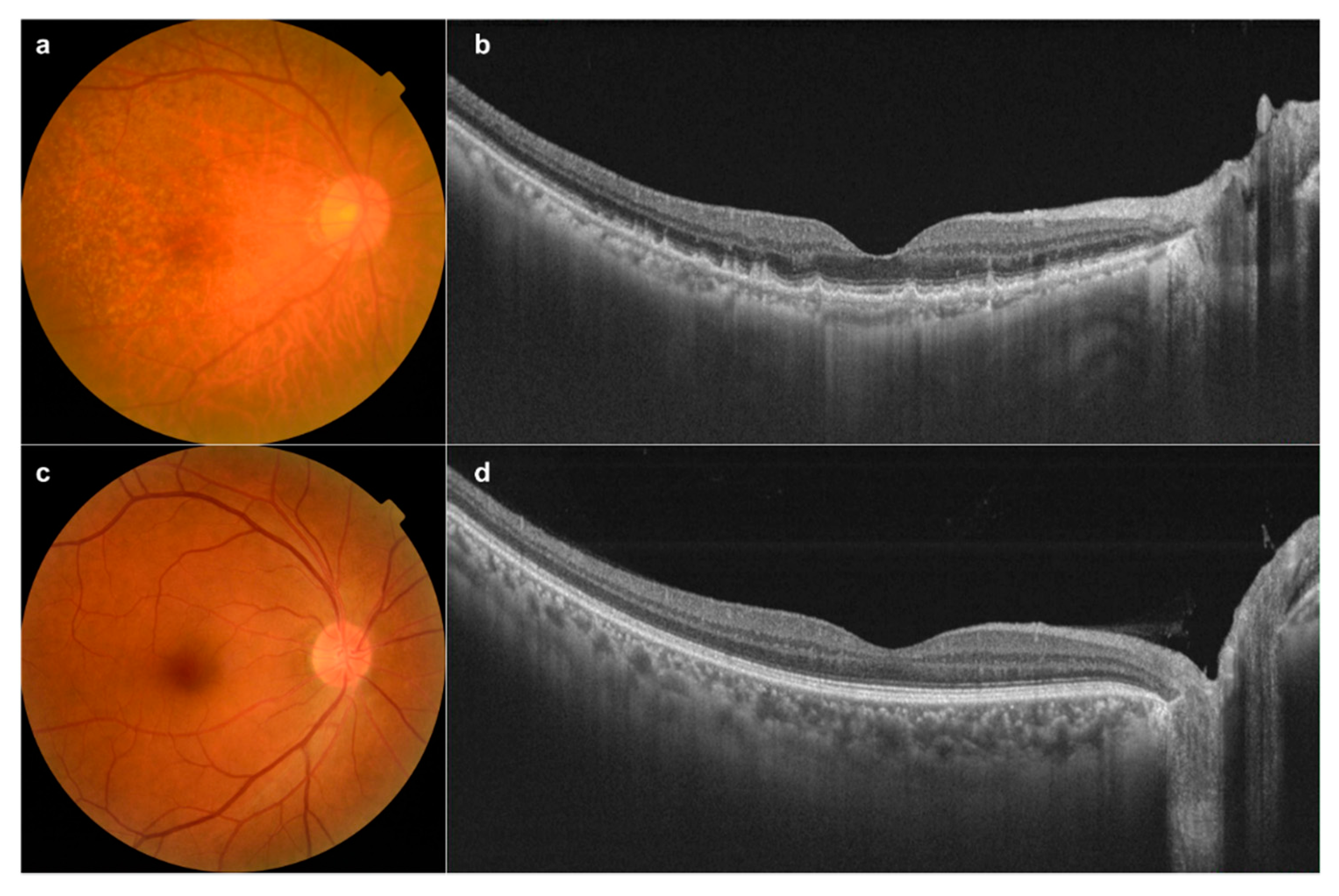
3.1. Fundus Image Collection
3.2. Data Augmentation
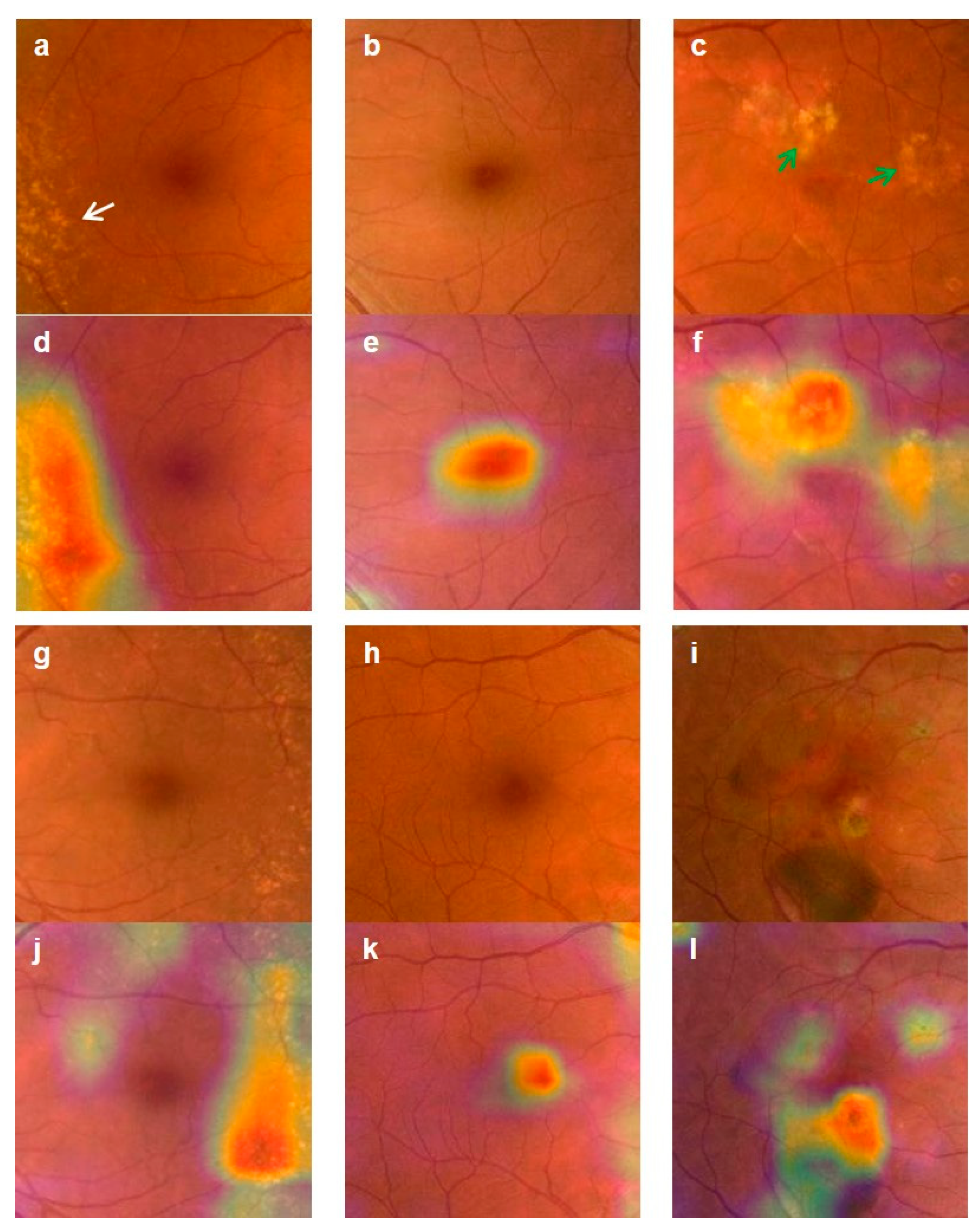
3.3. Validation of the Deep-Learning-Based Diagnostic Tool
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Levine, A.B.; Schlosser, C.; Grewal, J.; Coope, R.; Jones, S.J.M.; Yip, S. Rise of the Machines: Advances in Deep Learning for Cancer Diagnosis. Trends Cancer 2019, 5, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, R. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Mathews, S.M.; Kambhamettu, C.; Barner, K.E. A novel application of deep learning for single-lead ECG classification. Comput. Biol. Med. 2018, 99, 53–62. [Google Scholar] [CrossRef]
- Baltruschat, I.M.; Nickisch, H.; Grass, M.; Knopp, T.; Saalbach, A. Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification. Sci. Rep. 2019, 9, 6381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramoff, M.D.; Lavin, P.T.; Birch, M.; Shah, N.; Folk, J.C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit. Med. 2018, 1, 39. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Recognising and managing diabetic retinopathy. Commun. Eye Health 2011, 24, 5–9. [Google Scholar]
- Tremolada, G.; Del Turco, C.; Lattanzio, R.; Maestroni, S.; Maestroni, A.; Bandello, F.; Zerbini, G. The role of angiogenesis in the development of proliferative diabetic retinopathy: Impact of intravitreal anti-VEGF treatment. Exp. Diabetes Res. 2012, 2012, 728325. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.R. Risk factors for age-related macular degeneration. Prog. Retin. Eye Res. 2001, 20, 227–253. [Google Scholar] [CrossRef]
- Javadzadeh, A.; Ghorbanihaghjo, A.; Bahreini, E.; Rashtchizadeh, N.; Argani, H.; Alizadeh, S. Plasma oxidized LDL and thiol-containing molecules in patients with exudative age-related macular degeneration. Mol. Vis. 2010, 16, 2578–2584. [Google Scholar] [PubMed]
- Wang, J.J.; Rochtchina, E.; Lee, A.J.; Chia, E.M.; Smith, W.; Cumming, R.G.; Mitchell, P. Ten-year incidence and progression of age-related maculopathy: The blue Mountains Eye Study. Ophthalmology 2007, 114, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Cook, H.L.; Patel, P.J. Tufail A. Age-related macular degeneration: Diagnosis and management. Br. Med. Bull. 2008, 85, 127–149. [Google Scholar] [CrossRef] [PubMed]
- Talks, J.; Koshy, Z.; Chatzinikolas, K. Use of optical coherence tomography, fluorescein angiography and indocyanine green angiography in a screening clinic for wet age-related macular degeneration. Br. J. Ophthalmol. 2007, 91, 600–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardes, R.; Serranho, P.; Lobo, C. Digital ocular fundus imaging: A review. Ophthalmologica 2011, 226, 161–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition; Cornell University: Ithaca, NY, USA, 2014. [Google Scholar]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol. Retina 2017, 1, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Group A-REDSR. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.H.; Bhandary, S.V.; Sivaprasad, S.; Hagiwara, Y.; Bagchi, A.; Raghavendra, U.; Rao, A.K.; Raju, B.; Shetty, N.S.; Gertych, A.; et al. Age-related macular degeneration detection using deep convolutional neural network. Future Gener. Comput. Syst. 2018, 87, 127–135. [Google Scholar] [CrossRef]
- Ly, A.; Yapp, M.; Nivison-Smith, L.; Assaad, N.; Hennessy, M.; Kalloniatis, M. Developing prognostic biomarkers in intermediate age-related macular degeneration: Their clinical use in predicting progression. Clin. Exp. Optom. 2018, 101, 172–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handa, J.T.; Bowes Rickman, C.; Dick, A.D.; Gorin, M.B.; Miller, J.W.; Toth, C.A.; Ueffing, M.; Zarbin, M.; Farrer, L.A. A systems biology approach towards understanding and treating non-neovascular age-related macular degeneration. Nat. Commun. 2019, 10, 3347. [Google Scholar] [CrossRef] [PubMed]
- Campochiaro, P.A.; Marcus, D.M.; Awh, C.C.; Regillo, C.; Adamis, A.P.; Bantseev, V.; Chiang, Y.; Ehrlich, J.S.; Erickson, S.; Hanley, W.D.; et al. The Port Delivery System with Ranibizumab for Neovascular Age-Related Macular Degeneration: Results from the Randomized Phase 2 Ladder Clinical Trial. Ophthalmology 2019, 126, 1141–1154. [Google Scholar] [CrossRef] [PubMed]

| Average Accuracy | 3-Class | 2-Class | ||
|---|---|---|---|---|
| Control–dAMD–nAMD | Control–dAMD | Control–nAMD | dAMD–nAMD | |
| w-Pre | 0.9086 | 0.9192 | 0.9813 | 0.9132 |
| w/o-Pre | 0.8559 | 0.9264 | 0.9808 | 0.9063 |
| Folds | 3-Class | 2-Class | ||
|---|---|---|---|---|
| Normal–dAMD–nAMD | Normal–dAMD | Normal–nAMD | dAMD–nAMD | |
| Fold 1 | 0.9756 | 0.8846 | 1.0000 | 0.9231 |
| Fold 2 | 0.8864 | 1.0000 | 1.0000 | 0.8929 |
| Fold 3 | 0.9535 | 0.9259 | 1.0000 | 1.0000 |
| Fold 4 | 0.9318 | 0.9286 | 1.0000 | 0.9643 |
| Fold 5 | 0.7955 | 0.8571 | 0.9063 | 0.7857 |
| Average | 0.9086 | 0.9192 | 0.9813 | 0.9132 |
| Folds | 3-Class | 2-Class | ||
|---|---|---|---|---|
| Normal–dAMD–nAMD | Normal–dAMD | Normal–nAMD | dAMD–nAMD | |
| Fold 1 | 0.8049 | 0.8846 | 0.9667 | 0.9615 |
| Fold 2 | 0.8409 | 0.9286 | 1.0000 | 0.8571 |
| Fold 3 | 0.8837 | 0.9259 | 1.0000 | 0.9626 |
| Fold 4 | 0.8864 | 0.9643 | 1.0000 | 0.8214 |
| Fold 5 | 0.8636 | 0.9286 | 0.9375 | 0.9286 |
| Average | 0.8559 | 0.9264 | 0.9808 | 0.9063 |
| Model | Accuracy | Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|---|---|
| 3-class | 0.9086 | 0.9046 | 1.0000 | 1.0000 | 0.9349 | |
| Control–dAMD–nAMD | 0.8605 | 0.9394 | 0.8303 | 0.9500 | ||
| 0.9571 | 0.9329 | 0.8750 | 0.9786 | |||
| 2-class | Control–dAMD | 0.9192 | 0.9252 | 0.9167 | 0.8788 | 0.9492 |
| Control–nAMD | 0.9813 | 0.9684 | 1.0000 | 1.0000 | 0.9625 | |
| dAMD–nAMD | 0.9132 | 0.8795 | 0.9448 | 0.9318 | 0.8992 | |
| Folds | 3-Class | 2-Class | ||
|---|---|---|---|---|
| Normal–dAMD–nAMD | Normal–dAMD | Normal–nAMD | dAMD–nAMD | |
| Fold 1 | 0.7885 | 0.6071 | 0.8636 | 0.8636 |
| Fold 2 | 0.7885 | 0.7143 | 0.8182 | 0.7727 |
| Fold 3 | 0.6481 | 0.8571 | 0.9130 | 0.6957 |
| Fold 4 | 0.7500 | 0.7857 | 0.9545 | 0.7727 |
| Fold 5 | 0.6852 | 0.8276 | 1.0000 | 0.6957 |
| Average | 0.7321 | 0.7584 | 0.9099 | 0.7601 |
| Folds | 3-Class | 2-Class | ||||||
|---|---|---|---|---|---|---|---|---|
| Normal–dAMD–nAMD | Normal–dAMD | Normal–nAMD | dAMD–nAMD | |||||
| Reviewer 1 | Reviewer 2 | Reviewer 1 | Reviewer 2 | Reviewer 1 | Reviewer 2 | Reviewer 1 | Reviewer 2 | |
| Fold 1 | 0.7317 | 0.9024 | 0.9615 | 0.9231 | 0.9667 | 1.0000 | 0.6923 | 0.9231 |
| Fold 2 | 0.7045 | 0.9091 | 0.9643 | 0.8929 | 0.9375 | 0.9062 | 0.8519 | 0.9259 |
| Fold 3 | 0.6977 | 0.8140 | 0.8519 | 0.9259 | 0.9375 | 0.8750 | 0.8148 | 0.9630 |
| Fold 4 | 0.7955 | 0.7273 | 0.9643 | 0.9286 | 0.9062 | 0.9688 | 0.7500 | 0.9286 |
| Fold 5 | 0.7143 | 0.8000 | 0.7500 | 0.9643 | 0.8750 | 0.9062 | 0.7143 | 0.7143 |
| Average | 0.7287 | 0.8306 | 0.8984 | 0.9270 | 0.9246 | 0.9312 | 0.7647 | 0.8910 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heo, T.-Y.; Kim, K.M.; Min, H.K.; Gu, S.M.; Kim, J.H.; Yun, J.; Min, J.K. Development of a Deep-Learning-Based Artificial Intelligence Tool for Differential Diagnosis between Dry and Neovascular Age-Related Macular Degeneration. Diagnostics 2020, 10, 261. https://doi.org/10.3390/diagnostics10050261
Heo T-Y, Kim KM, Min HK, Gu SM, Kim JH, Yun J, Min JK. Development of a Deep-Learning-Based Artificial Intelligence Tool for Differential Diagnosis between Dry and Neovascular Age-Related Macular Degeneration. Diagnostics. 2020; 10(5):261. https://doi.org/10.3390/diagnostics10050261
Chicago/Turabian StyleHeo, Tae-Young, Kyoung Min Kim, Hyun Kyu Min, Sun Mi Gu, Jae Hyun Kim, Jaesuk Yun, and Jung Kee Min. 2020. "Development of a Deep-Learning-Based Artificial Intelligence Tool for Differential Diagnosis between Dry and Neovascular Age-Related Macular Degeneration" Diagnostics 10, no. 5: 261. https://doi.org/10.3390/diagnostics10050261
APA StyleHeo, T.-Y., Kim, K. M., Min, H. K., Gu, S. M., Kim, J. H., Yun, J., & Min, J. K. (2020). Development of a Deep-Learning-Based Artificial Intelligence Tool for Differential Diagnosis between Dry and Neovascular Age-Related Macular Degeneration. Diagnostics, 10(5), 261. https://doi.org/10.3390/diagnostics10050261





