KCTD15 Protein Expression in Peripheral Blood and Acute Myeloid Leukemia
Abstract
:1. Introduction
2. Results
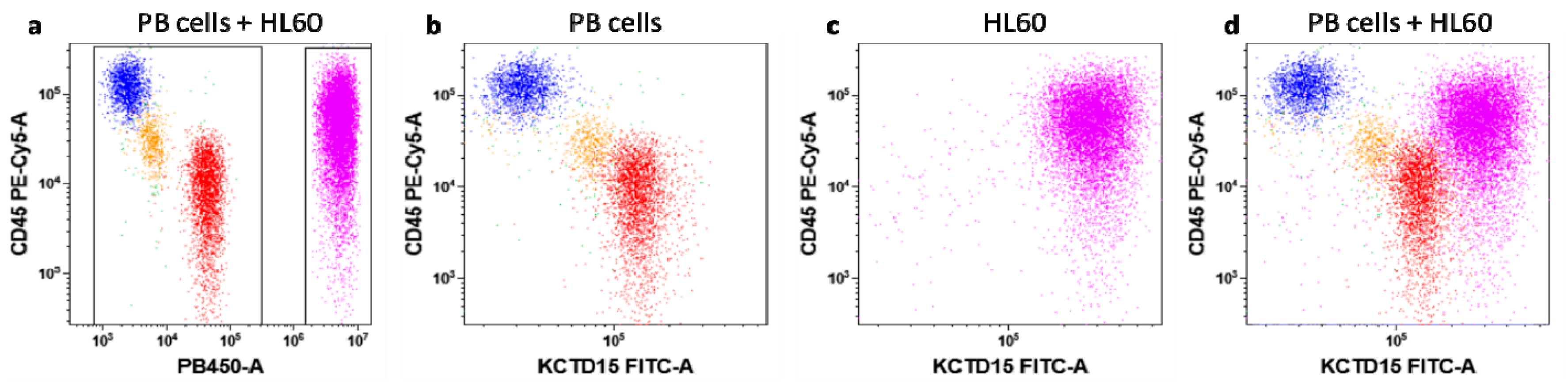
3.1. Intracellular KCTD15 Expression in Peripheral Blood
3.2. KCTD15 Protein Expression in Human Acute Myeloid Cell Lines
3.3. KCTD15 mRNA Expression in AML and Healthy BM
3. Discussion
4. Materials and Methods
4.1. Flow Cytometry Experiments
4.2. Western Blot Assays
4.3. Statistical Analysis and Reproducibility
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Pandya, P.H.; Murray, M.E.; Pollok, K.E.; Renbarger, J.L. The Immune System in Cancer Pathogenesis: Potential Therapeutic Approaches. J. Immunol. Res. 2016, 2016, 4273943. [Google Scholar] [CrossRef] [PubMed]
- Ivanovs, A.; Rybtsov, S.; Ng, E.S.; Stanley, E.G.; Elefanty, A.G.; Medvinsky, A. Human haematopoietic stem cell development: From the embryo to the dish. Development 2017, 144, 2323–2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurecic, R. Hematopoietic Stem Cell Heterogeneity. Adv. Exp. Med. Biol. 2019, 1169, 195–211. [Google Scholar] [CrossRef]
- Trowbridge, I.S.; Thomas, M.L. CD45: An emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu. Rev. Immunol. 1994, 12, 85–116. [Google Scholar] [CrossRef]
- Kirberg, J.; Brocker, T. CD45 up-regulation during lymphocyte maturation. Int. Immunol. 1996, 8, 1743–1749. [Google Scholar] [CrossRef]
- Poppema, S.; Lai, R.; Visser, L.; Yan, X.J. CD45 (leucocyte common antigen) expression in T and B lymphocyte subsets. Leuk. Lymphoma 1996, 20, 217–222. [Google Scholar] [CrossRef]
- Lambert, C.; Sack, U. Monocytes and macrophages in flow cytometry. Cytom. B Clin. Cytom. 2017, 92, 178–179. [Google Scholar] [CrossRef] [Green Version]
- Marimuthu, R.; Francis, H.; Dervish, S.; Li, S.C.H.; Medbury, H.; Williams, H. Characterization of Human Monocyte Subsets by Whole Blood Flow Cytometry Analysis. J. Vis. Exp. 2018. [Google Scholar] [CrossRef] [Green Version]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Arber, D.A. The 2016 WHO classification of acute myeloid leukemia: What the practicing clinician needs to know. Semin. Hematol. 2019, 56, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Skoblov, M.; Marakhonov, A.; Marakasova, E.; Guskova, A.; Chandhoke, V.; Birerdinc, A.; Baranova, A. Protein partners of KCTD proteins provide insights about their functional roles in cell differentiation and vertebrate development. Bioessays 2013, 35, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Aouacheria, A.; Lionnard, L.; Metz, K.A.; Soane, L.; Kamiya, A.; Hardwick, J.M. KCTD: A new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci. Ther. 2019, 25, 887–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smaldone, G.; Beneduce, G.; Incoronato, M.; Pane, K.; Franzese, M.; Coppola, L.; Cordella, A.; Parasole, R.; Ripaldi, M.; Nassa, G.; et al. KCTD15 is overexpressed in human childhood B-cell acute lymphoid leukemia. Sci. Rep. 2019, 9, 20108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagger, F.O.; Kinalis, S.; Rapin, N. BloodSpot: A database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 2019, 47, D881–D885. [Google Scholar] [CrossRef] [Green Version]
- Di Noto, R.; Mirabelli, P.; Del Vecchio, L. Flow cytometry analysis of acute promyelocytic leukemia: The power of ‘surface hematology’. Leukemia 2007, 21, 4–8. [Google Scholar] [CrossRef]
- D’Alessio, F.; Mirabelli, P.; Gorrese, M.; Scalia, G.; Gemei, M.; Mariotti, E.; Di Noto, R.; Martinelli, P.; Fortunato, G.; Paladini, D.; et al. Polychromatic flow cytometry analysis of CD34+ hematopoietic stem cells in cryopreserved early preterm human cord blood samples. Cytom. A 2011, 79, 14–24. [Google Scholar] [CrossRef]
- Bayón, Y.; Trinidad, A.G.; de la Puerta, M.L.; Del Carmen Rodríguez, M.; Bogetz, J.; Rojas, A.; De Pereda, J.M.; Rahmouni, S.; Williams, S.; Matsuzawa, S.; et al. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008, 275, 3900–3910. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, L.; Zhou, Y.; Shi, A.; Wang, H.; Han, M.; Wan, X.; Kilonzo, S.B.; Luo, X.; Chen, T.; et al. Interference with KCTD9 inhibits NK cell activation and ameliorates fulminant liver failure in mice. BMC Immunol. 2018, 19, 20. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef] [Green Version]
- Back, A.L.; Gollahon, K.A.; Hickstein, D.D. Regulation of expression of the leukocyte integrin CD11a (LFA-1) molecule during differentiation of HL-60 cells along the monocyte/macrophage pathway. J. Immunol. 1992, 148, 710–714. [Google Scholar] [PubMed]
- Idres, N.; Benoît, G.; Flexor, M.A.; Lanotte, M.; Chabot, G.G. Granulocytic differentiation of human NB4 promyelocytic leukemia cells induced by all-trans retinoic acid metabolites. Cancer Res. 2001, 61, 700–705. [Google Scholar] [PubMed]
- Bianchi, N.; Ongaro, F.; Chiarabelli, C.; Gualandi, L.; Mischiati, C.; Bergamini, P.; Gambari, R. Induction of erythroid differentiation of human K562 cells by cisplatin analogs. Biochem. Pharmacol. 2000, 60, 31–40. [Google Scholar] [CrossRef]
- Prada-Arismendy, J.; Arroyave, J.C.; Röthlisberger, S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017, 31, 63–76. [Google Scholar] [CrossRef]
- Kayser, S.; Levis, M.J. Clinical implications of molecular markers in acute myeloid leukemia. Eur. J. Haematol. 2019, 102, 20–35. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Xiang, Y.; Sun, G. The KCTD family of proteins: Structure, function, disease relevance. Cell Biosci. 2013, 3, 45. [Google Scholar] [CrossRef] [Green Version]
- Canettieri, G.; Di Marcotullio, L.; Greco, A.; Coni, S.; Antonucci, L.; Infante, P.; Pietrosanti, L.; De Smaele, E.; Ferretti, E.; Miele, E.; et al. Histone deacetylase and Cullin3-RENKCTD11 ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010, 12, U132–U191. [Google Scholar] [CrossRef]
- Moen, M.N.; Fjaer, R.; Hamdani, E.H.; Laerdahl, J.K.; Menchini, R.J.; Vigeland, M.D.; Sheng, Y.; Undlien, D.E.; Hassel, B.; Salih, M.A.; et al. Pathogenic variants in KCTD7 perturb neuronal K+ fluxes and glutamine transport. Brain 2016, 139, 3109–3120. [Google Scholar] [CrossRef] [Green Version]
- Golzio, C.; Willer, J.; Talkowski, M.E.; Oh, E.C.; Taniguchi, Y.; Jacquemont, S.; Reymond, A.; Sun, M.; Sawa, A.; Gusella, J.F.; et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 2012, 485, 363–367. [Google Scholar] [CrossRef]
- Marneros, A.G.; Beck, A.E.; Turner, E.H.; McMillin, M.J.; Edwards, M.J.; Field, M.; Sobreira, N.L.d.M.; Perez, A.B.A.; Fortes, J.A.R.; Lampe, A.K.; et al. Mutations in KCTD1 Cause Scalp-Ear-Nipple Syndrome. Am. J. Hum. Genet. 2013, 92, 621–626. [Google Scholar] [CrossRef] [Green Version]
- Smaldone, G.; Balasco, N.; Pirone, L.; Caruso, D.; Di Gaetano, S.; Pedone, E.M.; Vitagliano, L. Molecular basis of the scalp-ear-nipple syndrome unraveled by the characterization of disease-causing KCTD1 mutants. Sci. Rep. 2019, 9, 10519. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.J.; Goergen, P.; Rajendran, J.; Zheleznyakova, G.; Hagglund, M.G.; Perland, E.; Bagchi, S.; Kalogeropoulou, A.; Khan, Z.; Fredriksson, R.; et al. Obesity-Linked Homologues TfAP-2 and Twz Establish Meal Frequency in Drosophila melanogaster. PLoS Genet. 2014, 10, e1004499. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.; Pirone, L.; Capolupo, A.; Vitagliano, L.; Monti, M.C.; Di Gaetano, S.; Pedone, E. The essential player in adipogenesis GRP78 is a novel KCTD15 interactor. Inter. J. Biol. Macromol. 2018. [Google Scholar] [CrossRef]
- Pirone, L.; Smaldone, G.; Spinelli, R.; Barberisi, M.; Beguinot, F.; Vitagliano, L.; Miele, C.; Di Gaetano, S.; Raciti, G.A.; Pedone, E. KCTD1: A novel modulator of adipogenesis through the interaction with the transcription factor AP2α. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 158514. [Google Scholar] [CrossRef]
- Zarelli, V.E.; Dawid, I.B. Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proc. Natl. Acad. Sci. USA 2013, 110, 2870–2875. [Google Scholar] [CrossRef] [Green Version]
- Spiombi, E.; Angrisani, A.; Fonte, S.; De Feudis, G.; Fabretti, F.; Cucchi, D.; Izzo, M.; Infante, P.; Miele, E.; Po, A.; et al. KCTD15 inhibits the Hedgehog pathway in Medulloblastoma cells by increasing protein levels of the oncosuppressor KCASH2. Oncogenesis 2019, 8, 64. [Google Scholar] [CrossRef]
- Smaldone, G.; Pirone, L.; Balasco, N.; Di Gaetano, S.; Pedone, E.M.; Vitagliano, L. Cullin 3 Recognition Is Not a Universal Property among KCTD Proteins. PLoS ONE 2015, 10, e0126808. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.; Pirone, L.; Pedone, E.; Marlovits, T.; Vitagliano, L.; Ciccarelli, L. The BTB domains of the potassium channel tetramerization domain proteins prevalently assume pentameric states. Febs Lett. 2016, 590, 1663–1671. [Google Scholar] [CrossRef] [Green Version]
- Coppola, L.; Smaldone, G.; Cianflone, A.; Baselice, S.; Mirabelli, P.; Salvatore, M. Purification of viable peripheral blood mononuclear cells for biobanking using a robotized liquid handling workstation. J. Transl. Med. 2019, 17, 371. [Google Scholar] [CrossRef]
- Mirabelli, P.; Incoronato, M.; Coppola, L.; Infante, T.; Parente, C.A.; Nicolai, E.; Soricelli, A.; Salvatore, M. SDN Biobank: Bioresource of Human Samples Associated with Functional and/or Morphological Bioimaging Results for the Study of Oncological, Cardiological, Neurological, and Metabolic Diseases. Open J. Bioresour. 2017, 4, 2. [Google Scholar] [CrossRef]

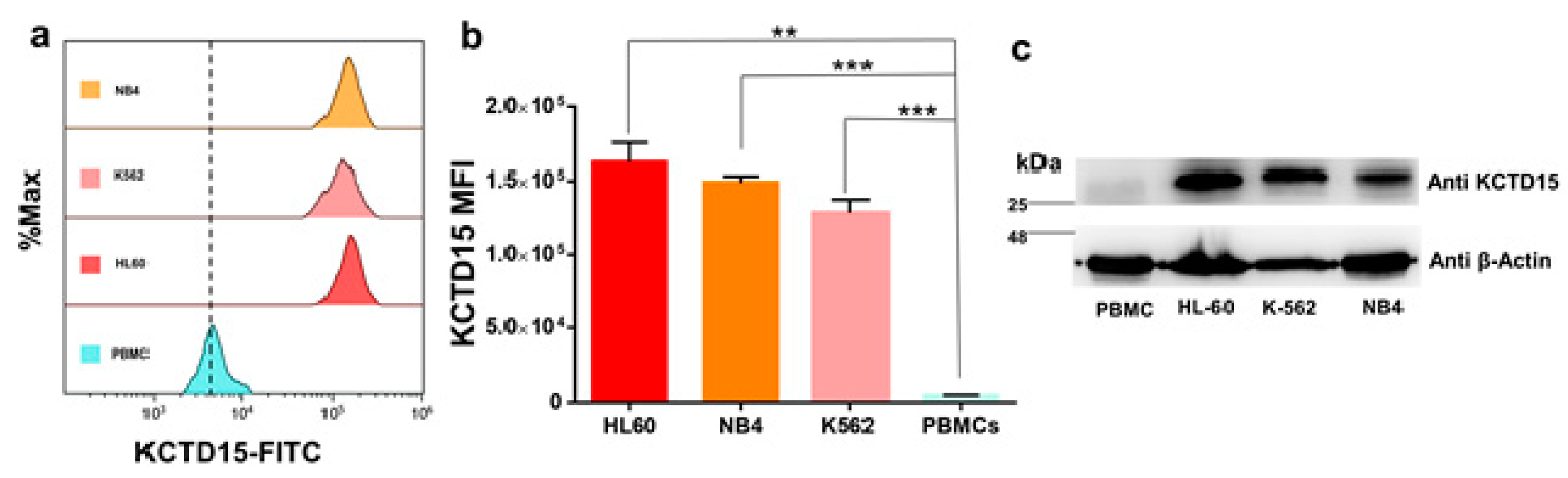

| HL-60 | NB-4 | K562 | PBMC | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| 162,296 | ±13,303 | 148,944 | ±4350 | 128,557 | ±8971 | 4622 | ±470.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smaldone, G.; Coppola, L.; Incoronato, M.; Parasole, R.; Ripaldi, M.; Vitagliano, L.; Mirabelli, P.; Salvatore, M. KCTD15 Protein Expression in Peripheral Blood and Acute Myeloid Leukemia. Diagnostics 2020, 10, 371. https://doi.org/10.3390/diagnostics10060371
Smaldone G, Coppola L, Incoronato M, Parasole R, Ripaldi M, Vitagliano L, Mirabelli P, Salvatore M. KCTD15 Protein Expression in Peripheral Blood and Acute Myeloid Leukemia. Diagnostics. 2020; 10(6):371. https://doi.org/10.3390/diagnostics10060371
Chicago/Turabian StyleSmaldone, Giovanni, Luigi Coppola, Mariarosaria Incoronato, Rosanna Parasole, Mimmo Ripaldi, Luigi Vitagliano, Peppino Mirabelli, and Marco Salvatore. 2020. "KCTD15 Protein Expression in Peripheral Blood and Acute Myeloid Leukemia" Diagnostics 10, no. 6: 371. https://doi.org/10.3390/diagnostics10060371
APA StyleSmaldone, G., Coppola, L., Incoronato, M., Parasole, R., Ripaldi, M., Vitagliano, L., Mirabelli, P., & Salvatore, M. (2020). KCTD15 Protein Expression in Peripheral Blood and Acute Myeloid Leukemia. Diagnostics, 10(6), 371. https://doi.org/10.3390/diagnostics10060371








