Development of a Nucleic Acid Lateral Flow Immunoassay for the Detection of Human Polyomavirus BK
Abstract
:1. Introduction
2. Materials and Methods
2.1. Target Area Detection and Probe Design
2.2. Preparation of AuNP–DP Conjugates
2.3. Fabrication of Lateral Flow Strip
2.4. Bacterial Culture and Nucleic Acid Extraction
2.5. Optimization, Sensitivity, and Specificity Assays
2.6. Assay Procedure
2.7. Statistical Analysis
3. Results
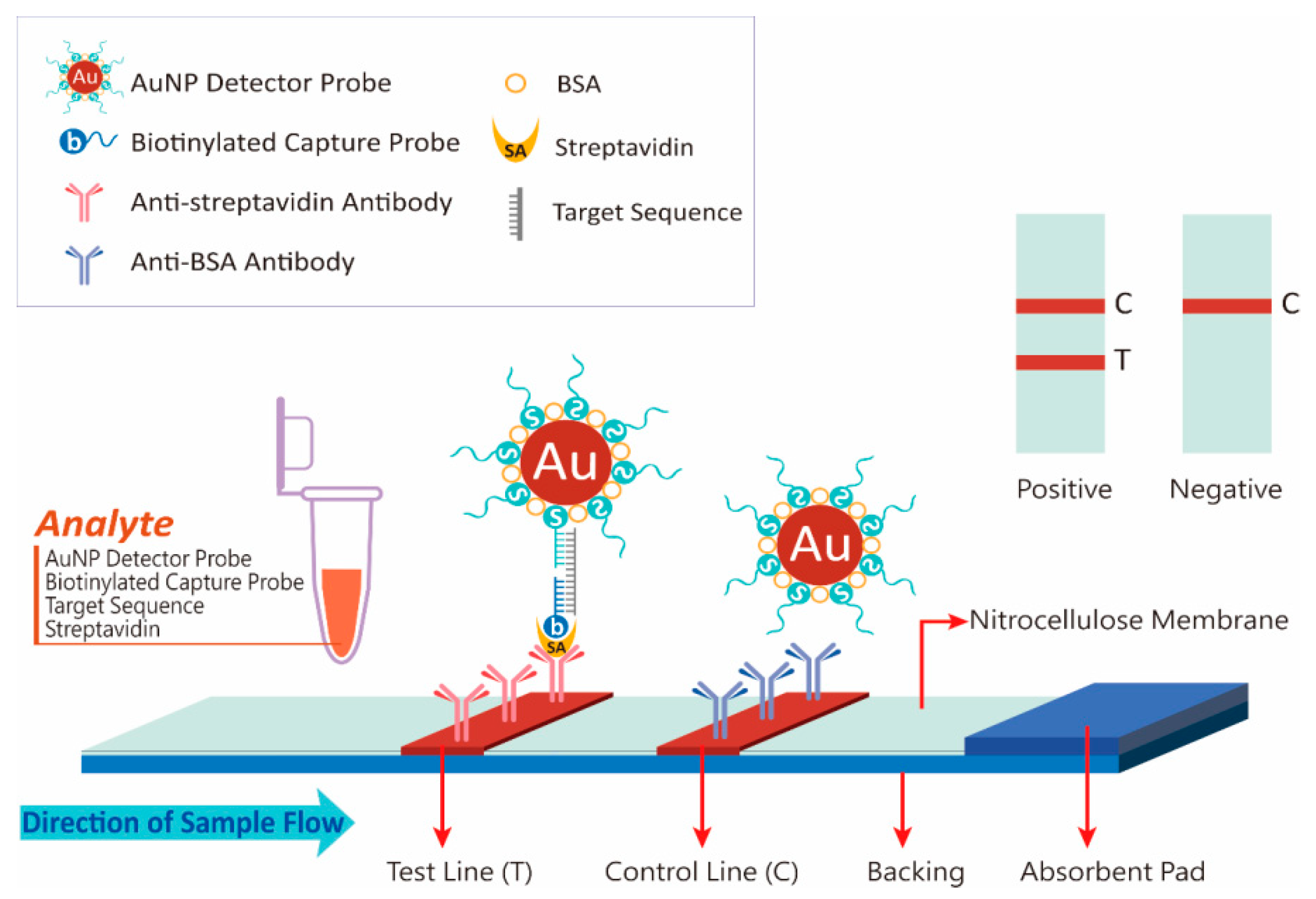
3.1. Principle of NALFIA Measurement and Probe Design
3.2. Conjugate Stability
3.3. Assay Optimization
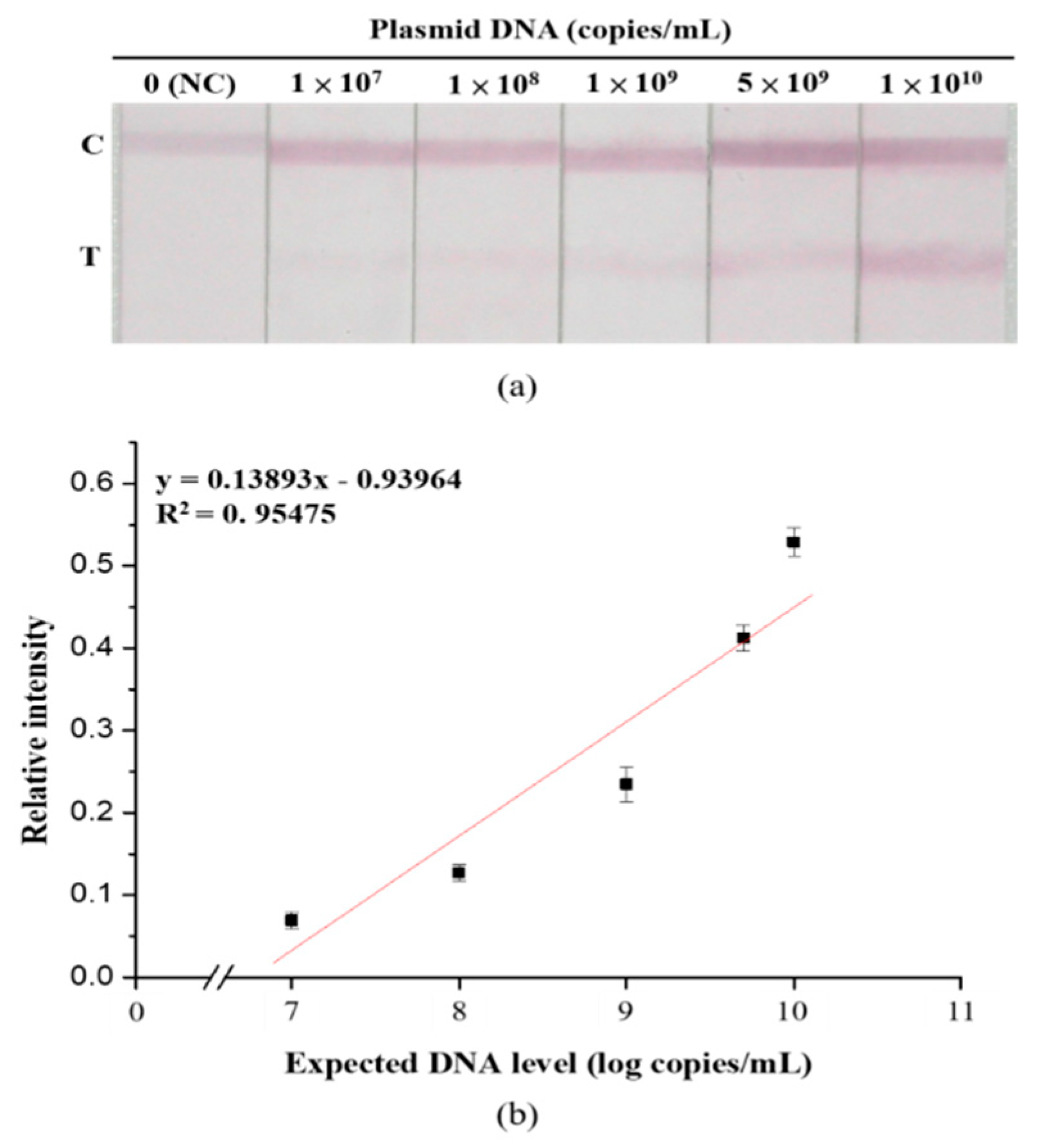
3.4. Sensitivity and Specificity Assays
3.5. Practicability of the Developed Lateral Flow Strip
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moens, U.; Calvignac-Spencer, S.; Lauber, C.; Ramqvist, T.; Feltkamp, M.C.W.; Daugherty, M.D.; Verschoor, E.J.; Ehlers, B.; Ictv Report, C. ICTV Virus taxonomy profile: Polyomaviridae. J. Gen. Virol. 2017, 98, 1159–1160. [Google Scholar] [CrossRef] [PubMed]
- Hurdiss, D.L.; Morgan, E.L.; Thompson, R.F.; Prescott, E.L.; Panou, M.M.; Macdonald, A.; Ranson, N.A. New structural insights into the genome and minor capsid proteins of BK polyomavirus using cryo-electron microscopy. Structure 2016, 24, 528–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolt, A.; Sasnauskas, K.; Koskela, P.; Lehtinen, M.; Dillner, J. Seroepidemiology of the human polyomaviruses. J. Gen. Virol. 2003, 84, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Knowles, W.A.; Pipkin, P.; Andrews, N.; Vyse, A.; Minor, P.; Brown, D.W.; Miller, E. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J. Med. Virol. 2003, 71, 115–123. [Google Scholar] [CrossRef]
- Gardner, S.D. Prevalence in England of antibody to human polyomavirus (B.k.). Br. Med. J. 1973, 1, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Shah, K.V.; Daniel, R.W.; Warszawski, R.M. High prevalence of antibodies to BK virus, an SV40-related papovavirus, in residents of Maryland. J. Infect. Dis. 1973, 128, 784–787. [Google Scholar] [CrossRef]
- Nickeleit, V.; Hirsch, H.H.; Binet, I.F.; Gudat, F.; Prince, O.; Dalquen, P.; Thiel, G.; Mihatsch, M.J. Polyomavirus infection of renal allograft recipients: From latent infection to manifest disease. J. Am. Soc. Nephrol. 1999, 10, 1080–1089. [Google Scholar]
- Boldorini, R.; Veggiani, C.; Barco, D.; Monga, G. Kidney and urinary tract polyomavirus infection and distribution: Molecular biology investigation of 10 consecutive autopsies. Arch. Pathol. Lab. Med. 2005, 129, 69–73. [Google Scholar]
- Rosen, S.; Harmon, W.; Krensky, A.M.; Edelson, P.J.; Padgett, B.L.; Grinnell, B.W.; Rubino, M.J.; Walker, D.L. Tubulo-interstitial nephritis associated with polyomavirus (BK type) infection. N. Engl. J. Med. 1983, 308, 1192–1196. [Google Scholar] [CrossRef]
- Coleman, D.V.; Mackenzie, E.F.; Gardner, S.D.; Poulding, J.M.; Amer, B.; Russell, W.J. Human polyomavirus (BK) infection and ureteric stenosis in renal allograft recipients. J. Clin. Pathol. 1978, 31, 338–347. [Google Scholar] [CrossRef] [Green Version]
- Arthur, R.R.; Shah, K.V.; Baust, S.J.; Santos, G.W.; Saral, R. Association of BK viruria with hemorrhagic cystitis in recipients of bone marrow transplants. N. Engl. J. Med. 1986, 315, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Nickeleit, V.; Singh, H.K. Polyomaviruses and disease: Is there more to know than viremia and viruria? Curr. Opin. Organ. Transplant. 2015, 20, 348–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binet, I.; Nickeleit, V.; Hirsch, H.H.; Prince, O.; Dalquen, P.; Gudat, F.; Mihatsch, M.J.; Thiel, G. Polyomavirus disease under new immunosuppressive drugs: A cause of renal graft dysfunction and graft loss. Transplantation 1999, 67, 918–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barraclough, K.A.; Isbel, N.M.; Staatz, C.E.; Johnson, D.W. BK Virus in kidney transplant recipients: The influence of immunosuppression. J. Transplant. 2011, 2011, 750836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickeleit, V.; Singh, H.K.; Mihatsch, M.J. Polyomavirus nephropathy: Morphology, pathophysiology, and clinical management. Curr. Opin. Nephrol. Hypertens. 2003, 12, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Sawinski, D.; Goral, S. BK virus infection: An update on diagnosis and treatment. Nephrol. Dial. Transplant. 2015, 30, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Hardinger, K.L.; Koch, M.J.; Bohl, D.J.; Storch, G.A.; Brennan, D.C. BK-virus and the impact of pre-emptive immunosuppression reduction: 5-year results. Am. J. Transplant. 2010, 10, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Comoli, P.; Cioni, M.; Basso, S.; Gagliardone, C.; Potenza, L.; Verrina, E.; Luppi, M.; Zecca, M.; Ghiggeri, G.M.; Ginevri, F. Immunity to polyomavirus BK infection: Immune monitoring to regulate the balance between risk of BKV nephropathy and induction of alloimmunity. Clin. Dev. Immunol. 2013, 2013, 256923. [Google Scholar] [CrossRef]
- Dekeyser, M.; Francois, H.; Beaudreuil, S.; Durrbach, A. Polyomavirus-specific cellular immunity: From BK-Virus-specific cellular immunity to BK-Virus-associated nephropathy? Front. Immunol. 2015, 6, 307. [Google Scholar] [CrossRef] [Green Version]
- Pavsic, J.; Devonshire, A.S.; Parkes, H.; Schimmel, H.; Foy, C.A.; Karczmarczyk, M.; Gutierrez-Aguirre, I.; Honeyborne, I.; Huggett, J.F.; McHugh, T.D.; et al. Standardization of nucleic acid tests for clinical measurements of bacteria and viruses. J. Clin. Microbiol. 2015, 53, 2008–2014. [Google Scholar] [CrossRef] [Green Version]
- Adam, B.; Randhawa, P.; Chan, S.; Zeng, G.; Regele, H.; Kushner, Y.B.; Colvin, R.B.; Reeve, J.; Mengel, M. Banff initiative for quality assurance in transplantation (BIFQUIT): Reproducibility of polyomavirus immunohistochemistry in kidney allografts. Am. J. Transplant. 2014, 14, 2137–2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab. Chip 2013, 13, 2210–2251. [Google Scholar] [CrossRef] [PubMed]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [PubMed]
- Liu, C.C.; Yeung, C.Y.; Chen, P.H.; Yeh, M.K.; Hou, S.Y. Salmonella detection using 16S ribosomal DNA/RNA probe-gold nanoparticles and lateral flow immunoassay. Food Chem. 2013, 141, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Somorin, Y.; O’Byrne, C. Determination of survival of wildtype and mutant Escherichia coli in soil. Bio-Protocol 2017, 7, e2414. [Google Scholar] [CrossRef] [Green Version]
- Goudsmit, J.; Wertheim-van Dillen, P.; van Strien, A.; van der Noordaa, J. The role of BK virus in acute respiratory tract disease and the presence of BKV DNA in tonsils. J. Med. Virol. 1982, 10, 91–99. [Google Scholar] [CrossRef]
- Sundsfjord, A.; Spein, A.R.; Lucht, E.; Flaegstad, T.; Seternes, O.M.; Traavik, T. Detection of BK virus DNA in nasopharyngeal aspirates from children with respiratory infections but not in saliva from immunodeficient and immunocompetent adult patients. J. Clin. Microbiol. 1994, 32, 1390–1394. [Google Scholar] [CrossRef] [Green Version]
- Raeesi, N.; Gheissari, A.; Akrami, M.; Moghim, S. Urinary BK virus excretion in children newly diagnosed with acute lymphoblastic leukemia. Int. J. Prev. Med. 2012, 3, 402–407. [Google Scholar]
- Vanchiere, J.A.; Nicome, R.K.; Greer, J.M.; Demmler, G.J.; Butel, J.S. Frequent detection of polyomaviruses in stool samples from hospitalized children. J. Infect. Dis. 2005, 192, 658–664. [Google Scholar] [CrossRef] [Green Version]
- Jeffers, L.K.; Madden, V.; Webster-Cyriaque, J. BK virus has tropism for human salivary gland cells in vitro: Implications for transmission. Virology 2009, 394, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, M.; Weyandt, T.B.; Frisque, R.J. Identification of archetype and rearranged forms of BK virus in leukocytes from healthy individuals. J. Med. Virol. 2000, 60, 353–362. [Google Scholar] [CrossRef]
- Rachmadi, A.T.; Torrey, J.R.; Kitajima, M. Human polyomavirus: Advantages and limitations as a human-specific viral marker in aquatic environments. Water Res. 2016, 105, 456–469. [Google Scholar] [CrossRef] [PubMed]
- NIAID Emerging Infectious Diseases/Pathogens. Available online: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed on 27 April 2020).
- Bofill-Mas, S.; Meschke, J.S.; Girones, R. Polyomavirus. Glob. Water Pathog. Proj. 2016. [Google Scholar] [CrossRef] [Green Version]
- Dalianis, T.; Hirsch, H.H. Human polyomaviruses in disease and cancer. Virology 2013, 437, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Engels, E.A.; Pfeiffer, R.M.; Fraumeni, J.F., Jr.; Kasiske, B.L.; Israni, A.K.; Snyder, J.J.; Wolfe, R.A.; Goodrich, N.P.; Bayakly, A.R.; Clarke, C.A.; et al. Spectrum of cancer risk among US solid organ transplant recipients. JAMA 2011, 306, 1891–1901. [Google Scholar] [CrossRef]
- Gupta, G.; Kuppachi, S.; Kalil, R.S.; Buck, C.B.; Lynch, C.F.; Engels, E.A. Treatment for presumed BK polyomavirus nephropathy and risk of urinary tract cancers among kidney transplant recipients in the United States. Am. J. Transplant. 2018, 18, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Bouvard, V.; Baan, R.A.; Grosse, Y.; Lauby-Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Straif, K. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 2012, 13, 339–340. [Google Scholar] [CrossRef]
- Hirsch, H.H.; Babel, N.; Comoli, P.; Friman, V.; Ginevri, F.; Jardine, A.; Lautenschlager, I.; Legendre, C.; Midtvedt, K.; Munoz, P.; et al. European perspective on human polyomavirus infection, replication and disease in solid organ transplantation. Clin. Microbiol. Infect. 2014, 20 (Suppl. 7), 74–88. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.G.; Knight, R.J.; Lunsford, K.E. BK virus as a mediator of graft dysfunction following kidney transplantation. Curr. Opin. Organ. Transplant. 2017, 22, 320–327. [Google Scholar] [CrossRef]
- Famulok, M.; Mayer, G. Chemical biology: Aptamers in nanoland. Nature 2006, 439, 666–669. [Google Scholar] [CrossRef]
- Storhoff, J.J.; Lazarides, A.A.; Mucic, R.C.; Mirkin, C.A.; Letsinger, R.L.; Schatz, G.C. What controls the optical properties of DNA-Linked gold nanoparticle assemblies? J. Am. Chem. Soc. 2000, 122, 4640–4650. [Google Scholar] [CrossRef]
- Liu, X.; Atwater, M.; Wang, J.; Huo, Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf. B Biointerfaces 2007, 58, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Nghiem, T.H.L.; La, T.H.; Vu, X.H.; Chu, V.H.; Nguyen, T.H.; Le, Q.H.; Fort, E.; Do, Q.H.; Tran, H.N. Synthesis, capping and binding of colloidal gold nanoparticles to proteins. Adv. Nat. Sci. Nanosci. Nanotechnol. 2010, 1, 025009. [Google Scholar] [CrossRef]
- Peterson, A.W.; Wolf, L.K.; Georgiadis, R.M. Hybridization of mismatched or partially matched DNA at surfaces. J. Am. Chem. Soc. 2002, 124, 14601–14607. [Google Scholar] [CrossRef] [PubMed]
- Randeria, P.S.; Jones, M.R.; Kohlstedt, K.L.; Banga, R.J.; Olvera de la Cruz, M.; Schatz, G.C.; Mirkin, C.A. What controls the hybridization thermodynamics of spherical nucleic acids? J. Am. Chem. Soc. 2015, 137, 3486–3489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demers, L.M.; Mirkin, C.A.; Mucic, R.C.; Reynolds, R.A., 3rd; Letsinger, R.L.; Elghanian, R.; Viswanadham, G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 2000, 72, 5535–5541. [Google Scholar] [CrossRef] [PubMed]
- Baldock, B.L.; Hutchison, J.E. UV-Visible spectroscopy-based quantification of unlabeled DNA bound to gold nanoparticles. Anal. Chem. 2016, 88, 12072–12080. [Google Scholar] [CrossRef]
- Frisque, R.J.; Bream, G.L.; Cannella, M.T. Human polyomavirus JC virus genome. J. Virol. 1984, 51, 458–469. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.C.; Wu, R. BK virus DNA: Complete nucleotide sequence of a human tumor virus. Science 1979, 206, 456–462. [Google Scholar] [CrossRef]
- Martini, F.; Corallini, A.; Balatti, V.; Sabbioni, S.; Pancaldi, C.; Tognon, M. Simian virus 40 in humans. Infect. Agent Cancer 2007, 2, 13. [Google Scholar] [CrossRef] [Green Version]
- Ambalathingal, G.R.; Francis, R.S.; Smyth, M.J.; Smith, C.; Khanna, R. BK Polyomavirus: Clinical aspects, immune regulation, and emerging therapies. Clin. Microbiol. Rev. 2017, 30, 503–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Name | Sequence (5’ to 3’) |
|---|---|
| Detector probe (5’S) | HS-A10-TATGTATGAATAGAGTCTTAGGT |
| Detector probe (3’S) | TATGTATGAATAGAGTCTTAGGT-A10-SH |
| Capture probe (5’B) | Biotin-A10-GAAAGGAAGGTAAGTTGTTAAG |
| Capture probe (3’B) | GAAAGGAAGGTAAGTTGTTAAG-A10-Biotin |
| BKV target DNA | ATCTTAACAACTTACCTTCCTTTCCGACCTAAGACT CTATTCATACATAT |
| JCV target DNA | CTCCTAACACGTCACCTTTCTTTCCGACCTAAATCT TTATTCGTACATAT |
| SV40 target DNA | GTCTTAACACCTCACCTTTCTCTCTAACCTGTTTCT CAAATCAAACAGTC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-H.; Yu, K.-Y.; Huang, S.-P.; Chuang, H.-W.; Lin, W.-Z.; Cherng, J.-H.; Hung, Y.-W.; Yeh, M.-K.; Hong, P.-D.; Liu, C.-C. Development of a Nucleic Acid Lateral Flow Immunoassay for the Detection of Human Polyomavirus BK. Diagnostics 2020, 10, 403. https://doi.org/10.3390/diagnostics10060403
Huang Y-H, Yu K-Y, Huang S-P, Chuang H-W, Lin W-Z, Cherng J-H, Hung Y-W, Yeh M-K, Hong P-D, Liu C-C. Development of a Nucleic Acid Lateral Flow Immunoassay for the Detection of Human Polyomavirus BK. Diagnostics. 2020; 10(6):403. https://doi.org/10.3390/diagnostics10060403
Chicago/Turabian StyleHuang, Yi-Huei, Kuan-Yi Yu, Shou-Ping Huang, Hui-Wen Chuang, Wen-Zhi Lin, Juin-Hong Cherng, Yao-Wen Hung, Ming-Kung Yeh, Po-Da Hong, and Cheng-Che Liu. 2020. "Development of a Nucleic Acid Lateral Flow Immunoassay for the Detection of Human Polyomavirus BK" Diagnostics 10, no. 6: 403. https://doi.org/10.3390/diagnostics10060403
APA StyleHuang, Y.-H., Yu, K.-Y., Huang, S.-P., Chuang, H.-W., Lin, W.-Z., Cherng, J.-H., Hung, Y.-W., Yeh, M.-K., Hong, P.-D., & Liu, C.-C. (2020). Development of a Nucleic Acid Lateral Flow Immunoassay for the Detection of Human Polyomavirus BK. Diagnostics, 10(6), 403. https://doi.org/10.3390/diagnostics10060403





