Electrochemical Aptasensors: Current Status and Future Perspectives
Abstract
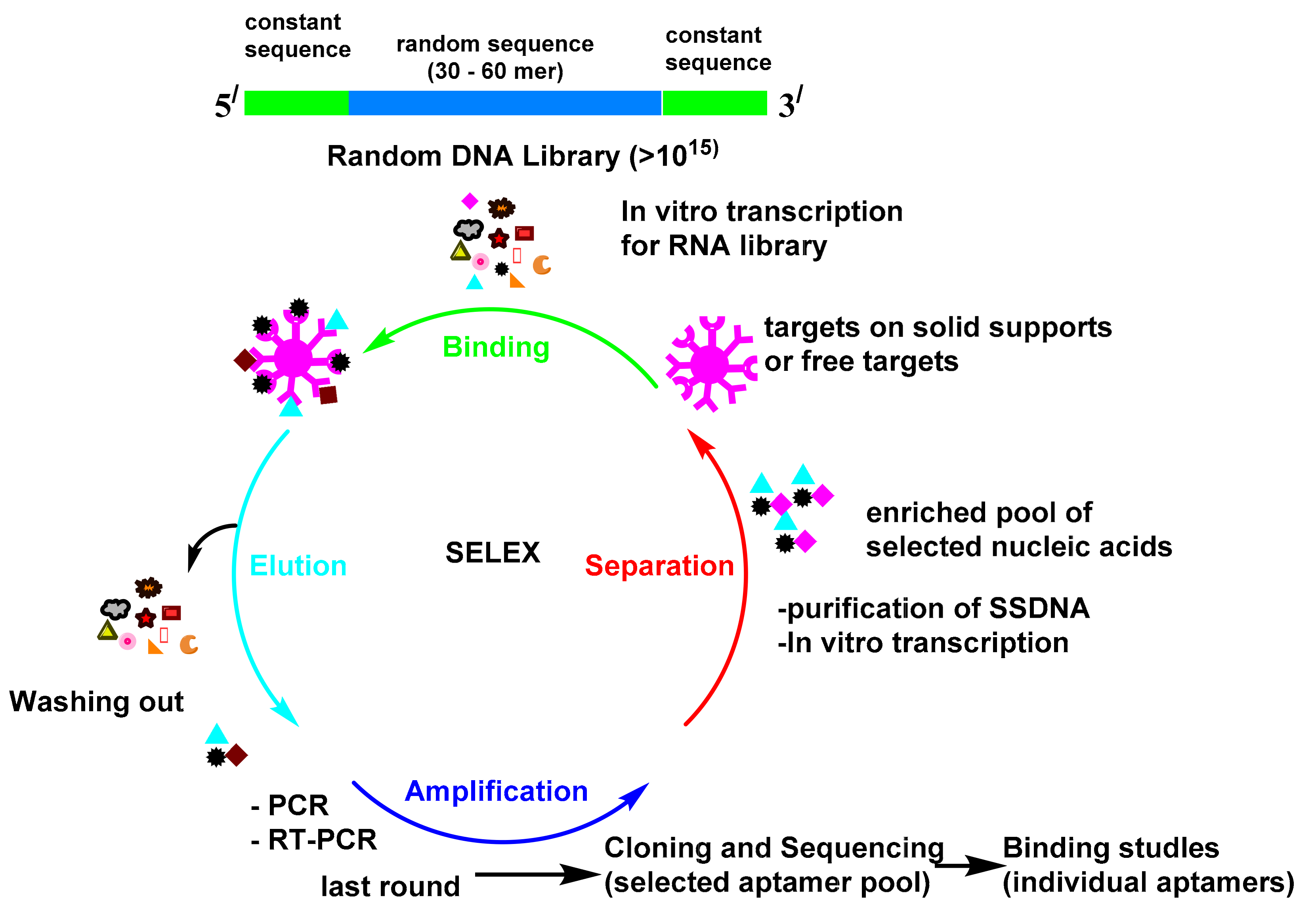
:1. Introduction
2. Immobilization Chemistry of Aptamer onto the Electrode Surface
3. Electrochemical Aptasensors Configuration
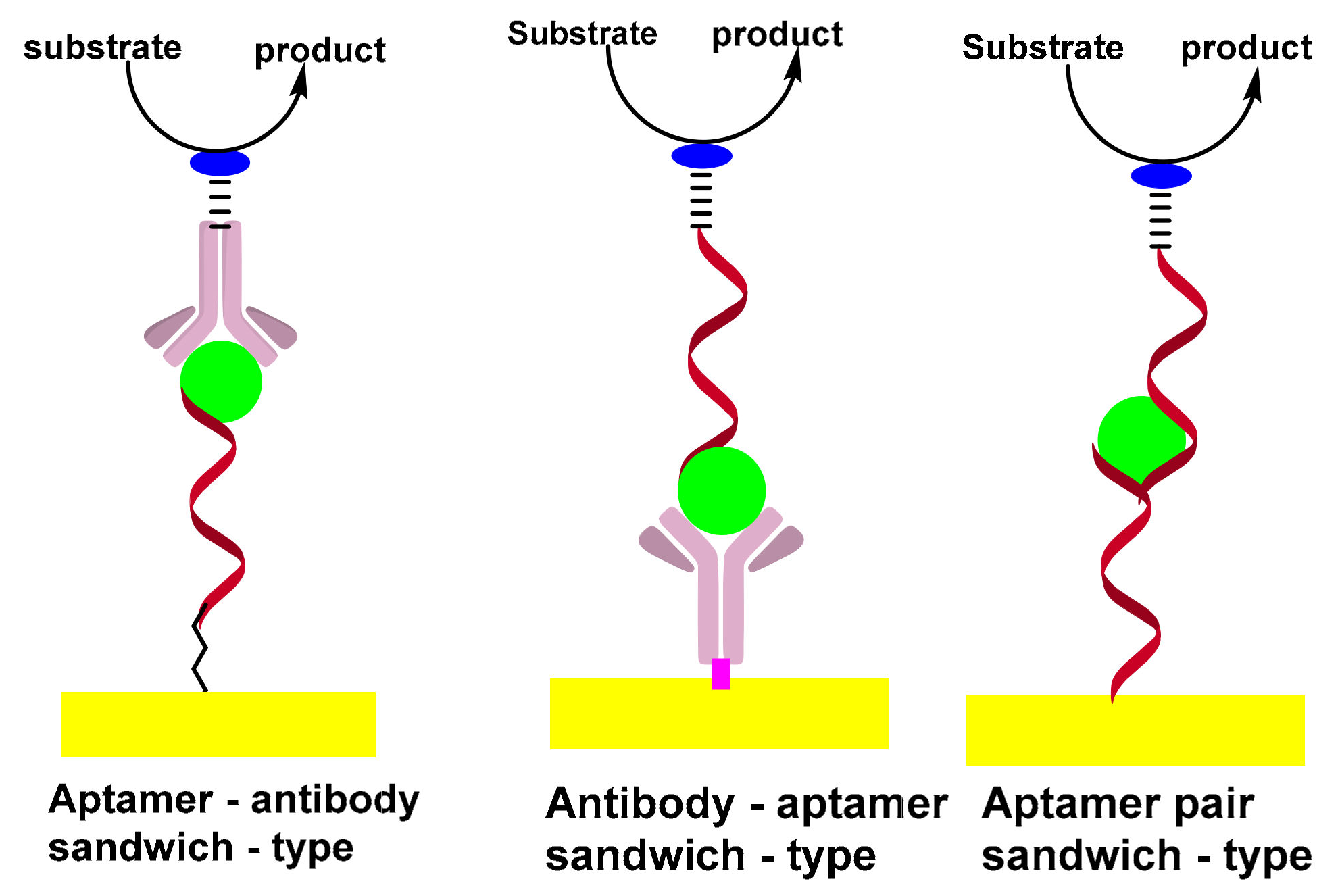
3.1. Sandwich-Type Electrochemical Aptasensors
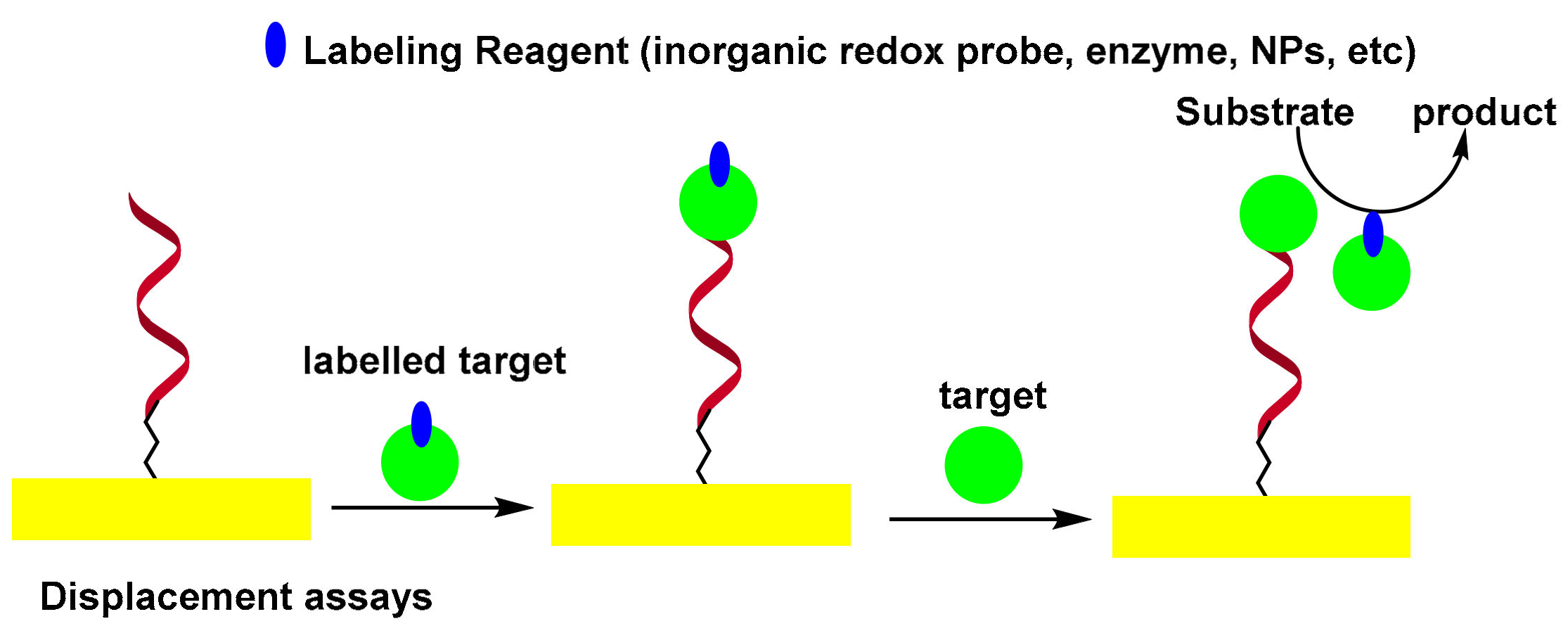
3.2. Displacement-Type Electrochemical Aptasensors
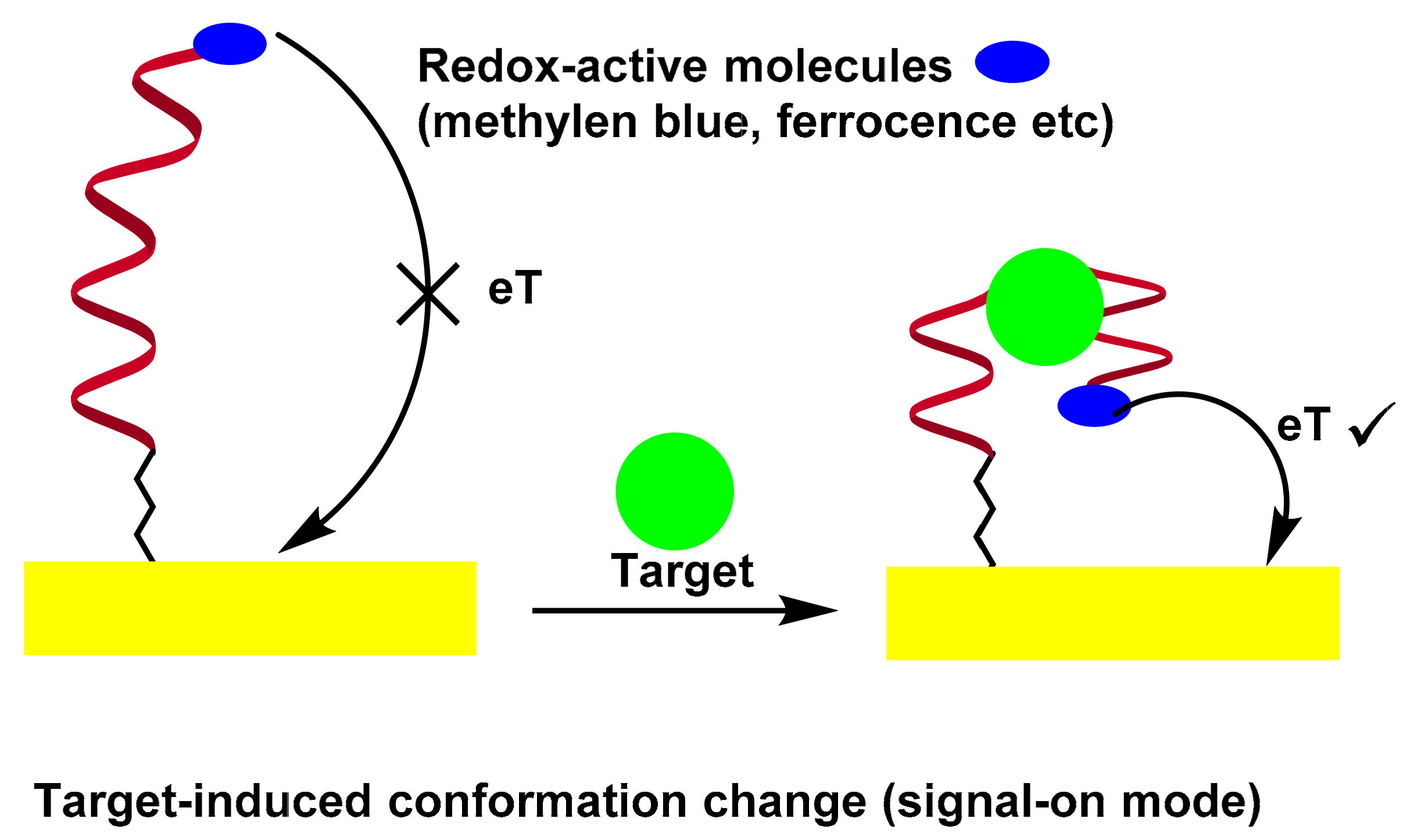
3.3. Folding-Based Electrochemical Aptasensor
4. Principles of Electrochemical Transduction Techniques
5. Electrochemical Aptasensors for Clinical Application
5.1. Electrochemical Aptasensors for Cardiac Biomarkers
5.2. Electrochemical Aptasensors for Alzheimer’s Disease (AD) Biomarkers
5.3. Electrochemical Aptasensors for Infectious Disease Biomarkers
5.4. Electrochemical Aptasensors for Cancer Diagnosis
5.4.1. Electrochemical Aptasensors for Circulating Tumor Cells (CTCs)
5.4.2. Electrochemical Aptasensors for Carcinoembryonic Antigens (CEAs)
5.4.3. Electrochemical Aptasensors for Exosomes
5.4.4. Electrochemical Aptasensors for Circulating Tumor Cells Biomarkers
6. Conclusions and Outlook
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Syed, M.A.; Pervaiz, S. Advances in Aptamers. Oligonucleotides 2010, 20, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current advances in aptamers for cancer diagnosis and therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Birch, J.R.; Racher, A.J. Antibody production. Adv. Drug Deliv. Rev. 2006, 58, 671–685. [Google Scholar] [CrossRef]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Mascini, M. Aptamers and their applications. Anal. Bioanal. Chem. 2008, 390, 987–988. [Google Scholar] [CrossRef]
- Ferreira, C.S.; Missailidis, S. Aptamer-based therapeutics and their potential in radiopharmaceutical design. Braz. Arch. Biol. Technol. 2007, 50, 63–76. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Han, K.; Liang, Z.; Zhou, N. Design strategies for aptamer-based biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef] [Green Version]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Electrochemical detection of protein using a double aptamer sandwich. Anal. Lett. 2004, 37, 2901–2909. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 2005, 20, 2168–2172. [Google Scholar] [CrossRef] [PubMed]
- Kara, P.; de la Escosura-Muñiz, A.; Maltez-da Costa, M.; Guix, M.; Ozsoz, M.; Merkoçi, A. Aptamers based electrochemical biosensor for protein detection using carbon nanotubes platforms. Biosens. Bioelectron. 2010, 26, 1715–1718. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhu, N.; Yu, P.; Mao, L. Aptamer-based electrochemical sensors that are not based on the target binding-induced conformational change of aptamers. Analyst 2008, 133, 1256–1260. [Google Scholar] [CrossRef]
- Yan, Z.; Han, Z.; Huang, H.; Shen, H.; Lu, X. Rational design of a thrombin electrochemical aptasensor by conjugating two DNA aptamers with G-quadruplex halves. Anal. Biochem. 2013, 442, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Lin, L.; Cheng, G.; Wang, A.; Tan, X.; He, P.; Fang, Y. Study on an electrochemical biosensor for thrombin recognition based on aptamers and nano particles. Sci. China Ser. B Chem. 2007, 50, 351–357. [Google Scholar] [CrossRef]
- Xu, D.; Xu, D.; Yu, X.; Liu, Z.; He, W.; Ma, Z. Label-Free Electrochemical Detection for Aptamer-Based Array Electrodes. Anal. Chem. 2005, 77, 5107–5113. [Google Scholar] [CrossRef]
- Numnuam, A.; Chumbimuni-Torres, K.Y.; Xiang, Y.; Bash, R.; Thavarungkul, P.; Kanatharana, P.; Pretsch, E.; Wang, J.; Bakker, E. Aptamer-Based Potentiometric Measurements of Proteins Using Ion-Selective Microelectrodes. Anal. Chem. 2008, 80, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, J.; Yun, W.; Xiao, S.; Chang, Z.; He, P.; Fang, Y. Detection of thrombin using electrogenerated chemiluminescence based on Ru(bpy)32+-doped silica nanoparticle aptasensor via target protein-induced strand displacement. Anal. Chim. Acta 2007, 598, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Sun, C.; Kang, Y.; Chen, J.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Label-free electrochemical detection of nanomolar adenosine based on target-induced aptamer displacement. Electrochem. Commun. 2008, 10, 531–535. [Google Scholar] [CrossRef]
- Cho, E.J.; Lee, J.-W.; Ellington, A.D. Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef] [PubMed]
- Leff, D.V.; Brandt, L.; Heath, J.R. Synthesis and characterization of hydrophobic, organically-soluble gold nanocrystals functionalized with primary amines. Langmuir 1996, 12, 4723–4730. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Gong, P.; Harbers, G.M.; Grainger, D.W.; Castner, D.G.; Gamble, L.J. Surface coverage and structure of mixed DNA/alkylthiol monolayers on gold: Characterization by XPS, NEXAFS, and fluorescence intensity measurements. Anal. Chem. 2006, 78, 3316–3325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keighley, S.D.; Li, P.; Estrela, P.; Migliorato, P. Optimization of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2008, 23, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Makaraviciute, A.; Kumar, S.; Wen, C.; Sjoödin, M.; Abdurakhmanov, E.; Danielson, U.H.; Nyholm, L.; Zhang, Z. Structural Changes of Mercaptohexanol Self-Assembled Monolayers on Gold and Their Influence on Impedimetric Aptamer Sensors. Anal. Chem. 2019, 91, 14697–14704. [Google Scholar] [CrossRef]
- Vericat, C.; Vela, M.; Benitez, G.; Carro, P.; Salvarezza, R. Self-assembled monolayers of thiols and dithiols on gold: New challenges for a well-known system. Chem. Soc. Rev. 2010, 39, 1805–1834. [Google Scholar] [CrossRef]
- Ying, G.; Wang, M.; Yi, Y.; Chen, J.; Mei, J.; Zhang, Y.; Chen, S. Construction and application of an electrochemical biosensor based on an endotoxin aptamer. Biotechnol. Appl. Biochem. 2018, 65, 323–327. [Google Scholar] [CrossRef]
- Porter, M.D.; Bright, T.B.; Allara, D.L.; Chidsey, C.E. Spontaneously organized molecular assemblies. 4. Structural characterization of n-alkyl thiol monolayers on gold by optical ellipsometry, infrared spectroscopy, and electrochemistry. J. Am. Chem. Soc. 1987, 109, 3559–3568. [Google Scholar] [CrossRef]
- Josephs, E.A.; Ye, T. A single-molecule view of conformational switching of DNA tethered to a gold electrode. J. Am. Chem. Soc. 2012, 134, 10021–10030. [Google Scholar] [CrossRef]
- Hanssen, B.L.; Siraj, S.; Wong, D.K. Recent strategies to minimise fouling in electrochemical detection systems. Rev. Anal. Chem. 2016, 35, 1–28. [Google Scholar] [CrossRef]
- Greenberg, A.; Breneman, C.M.; Liebman, J.F. The Amide Linkage: Structural Significance in Chemistry, Biochemistry, and Materials Science; John Wiley & Sons: New York, NY, USA, 2000. [Google Scholar]
- Rabai, S.; Benounis, M.; Catanante, G.; Baraket, A.; Errachid, A.; Jaffrezic Renault, N.; Marty, J.L.; Rhouati, A. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: Application to cadmium detection in water. Anal. Biochem. 2021, 612, 113956. [Google Scholar] [CrossRef] [PubMed]
- Gökçe, G.; Ben Aissa, S.; Nemčeková, K.; Catanante, G.; Raouafi, N.; Marty, J.L. Aptamer-modified pencil graphite electrodes for the impedimetric determination of ochratoxin A. Food Control 2020, 115, 107271. [Google Scholar] [CrossRef]
- Haghshenas, E.; Madrakian, T.; Afkhami, A.; Saify Nabiabad, H. An electrochemical ceruloplasmin aptasensor using a glassy carbon electrode modified by diazonium-functionalized multiwalled carbon nanotubes. J. Iran. Chem. Soc. 2019, 16, 593–602. [Google Scholar] [CrossRef]
- Khan, R.; Aissa, S.B.; Sherazi, T.A.; Catanante, G.; Hayat, A.; Marty, J.L. Development of an impedimetric aptasensor for label free detection of patulin in apple juice. Molecules 2019, 24, 1017. [Google Scholar] [CrossRef] [Green Version]
- Feier, B.; Băjan, I.; Cristea, C.; Săndulescu, R. Aptamer-based electrochemical sensor for the detection of ampicillin. In Proceedings of the International Conference on Advancements of Medicine and Health Care through Technology, Cluj-Napoca, Romania, 12–15 October 2016; pp. 107–110. [Google Scholar]
- Istamboulié, G.; Paniel, N.; Zara, L.; Granados, L.R.; Barthelmebs, L.; Noguer, T. Development of an impedimetric aptasensor for the determination of aflatoxin M1 in milk. Talanta 2016, 146, 464–469. [Google Scholar] [CrossRef]
- Belanger, D.; Pinson, J. Electrografting: A powerful method for surface modification. Chem. Soc. Rev. 2011, 40, 3995–4048. [Google Scholar] [CrossRef]
- Nwe, K.; Brechbiel, M.W. Growing applications of “click chemistry” for bioconjugation in contemporary biomedical research. Cancer Biother. Radiopharm. 2009, 24, 289–302. [Google Scholar] [CrossRef]
- Grabowska, I.; Sharma, N.; Vasilescu, A.; Iancu, M.; Badea, G.; Boukherroub, R.; Ogale, S.; Szunerits, S. Electrochemical Aptamer-Based Biosensors for the Detection of Cardiac Biomarkers. ACS Omega 2018, 3, 12010–12018. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.; Li, C.; Shangguan, L.; Qi, H.; Xue, D.; Gao, Q.; Zhang, C. Click chemistry-assisted self-assembly of DNA aptamer on gold nanoparticles-modified screen-printed carbon electrodes for label-free electrochemical aptasensor. Sens. Actuators B Chem. 2014, 192, 558–564. [Google Scholar] [CrossRef]
- Hayat, A.; Sassolas, A.; Marty, J.L.; Radi, A.E. Highly sensitive ochratoxin A impedimetric aptasensor based on the immobilization of azido-aptamer onto electrografted binary film via click chemistry. Talanta 2013, 103, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Zhang, Z.; Weng, W.; Wu, B.; Cheng, J.; Shi, L.; Sun, H.; Gao, L.; Shi, K. Highly sensitive detection of carcinoembryonic antigen using copper-free click chemistry on the surface of azide cofunctionalized graphene oxide. Anal. Chim. Acta 2020, 1127, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Tang, Y.; Yang, W.; Yu, Y. Disposable multiplexed electrochemical sensors based on electro-triggered selective immobilization of probes for simultaneous detection of DNA and proteins. J. Mater. Chem. B 2020, 8, 7501–7510. [Google Scholar] [CrossRef] [PubMed]
- Maehashi, K.; Matsumoto, K. Label-Free Electrical Detection Using Carbon Nanotube-Based Biosensors. Sensors 2009, 9, 5368–5378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hianik, T.; Ostatná, V.; Sonlajtnerova, M.; Grman, I. Influence of ionic strength, pH and aptamer configuration for binding affinity to thrombin. Bioelectrochemistry 2007, 70, 127–133. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, W.; Wang, J.; Yang, C.; Yang, F.; Yang, X. A sensitive impedimetric thrombin aptasensor based on polyamidoamine dendrimer. Talanta 2009, 78, 1240–1245. [Google Scholar] [CrossRef]
- Mir, M.; Vreeke, M.; Katakis, I. Different strategies to develop an electrochemical thrombin aptasensor. Electrochem. Commun. 2006, 8, 505–511. [Google Scholar] [CrossRef]
- Seo, H.B.; Gu, M.B. Aptamer-based sandwich-type biosensors. J. Biol. Eng. 2017, 11, 11. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Park, J.-W.; Kim, I.-A.; Youn, B.-S.; Gu, M.B. Sensitive detection of adipokines for early diagnosis of type 2 diabetes using enzyme-linked antibody-aptamer sandwich (ELAAS) assays. Sens. Actuators B Chem. 2012, 168, 243–248. [Google Scholar] [CrossRef]
- Shen, C.; Liu, S.; Li, X.; Yang, M. Electrochemical Detection of Circulating Tumor Cells Based on DNA Generated Electrochemical Current and Rolling Circle Amplification. Anal. Chem. 2019, 91, 11614–11619. [Google Scholar] [CrossRef]
- Sun, D.; Lu, J.; Luo, Z.; Zhang, L.; Liu, P.; Chen, Z. Competitive electrochemical platform for ultrasensitive cytosensing of liver cancer cells by using nanotetrahedra structure with rolling circle amplification. Biosens. Bioelectron. 2018, 120, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Tang, Y. Gold Nanoparticles-Based Multipedal DNA Walker for Ratiometric Detection of Circulating Tumor Cell. Anal. Chem. 2019, 91, 15187–15192. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, E.; Acero, J.L.; Reekmans, G.; Laureyn, W.; O’Sullivan, C.K. Displacement enzyme linked aptamer assay. Anal. Chem. 2005, 77, 4774–4784. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.; Jenkins, A.T.A.; Katakis, I. Ultrasensitive detection based on an aptamer beacon electron transfer chain. Electrochem. Commun. 2008, 10, 1533–1536. [Google Scholar] [CrossRef]
- Radi, A.E.; O’Sullivan, C.K. Aptamer conformational switch as sensitive electrochemical biosensor for potassium ion recognition. Chem. Commun. 2006, 3432–3434. [Google Scholar] [CrossRef]
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heeger, A.J. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991. [Google Scholar] [CrossRef]
- Sánchez, J.L.A.; Baldrich, E.; Radi, A.E.G.; Dondapati, S.; Sánchez, P.L.; Katakis, I.; O’Sullivan, C.K. Electronic ‘off-on’ molecular switch for rapid detection of thrombin. Electroanalysis 2006, 18, 1957–1962. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Huang, Y.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Electrochemical aptasensor based on proximity-dependent surface hybridization assay for single-step, reusable, sensitive protein detection. J. Am. Chem. Soc. 2007, 129, 15448–15449. [Google Scholar] [CrossRef]
- Kawde, A.-N.; Rodriguez, M.C.; Lee, T.M.; Wang, J. Label-free bioelectronic detection of aptamer–protein interactions. Electrochem. Commun. 2005, 7, 537–540. [Google Scholar] [CrossRef]
- Dou, B.; Xu, L.; Jiang, B.; Yuan, R.; Xiang, Y. Aptamer-Functionalized and Gold Nanoparticle Array-Decorated Magnetic Graphene Nanosheets Enable Multiplexed and Sensitive Electrochemical Detection of Rare Circulating Tumor Cells in Whole Blood. Anal. Chem. 2019, 91, 10792–10799. [Google Scholar] [CrossRef]
- Ou, D.; Sun, D.; Liang, Z.; Chen, B.; Lin, X.; Chen, Z. A novel cytosensor for capture, detection and release of breast cancer cells based on metal organic framework PCN-224 and DNA tetrahedron linked dual-aptamer. Sens. Actuators B Chem. 2019, 285, 398–404. [Google Scholar] [CrossRef]
- Tian, L.; Qi, J.; Qian, K.; Oderinde, O.; Liu, Q.; Yao, C.; Song, W.; Wang, Y. Copper (II) oxide nanozyme based electrochemical cytosensor for high sensitive detection of circulating tumor cells in breast cancer. J. Electroanal. Chem. 2018, 812, 1–9. [Google Scholar] [CrossRef]
- Liu, J.-X.; Bao, N.; Luo, X.; Ding, S.-N. Nonenzymatic Amperometric Aptamer Cytosensor for Ultrasensitive Detection of Circulating Tumor Cells and Dynamic Evaluation of Cell Surface N-Glycan Expression. ACS Omega 2018, 3, 8595–8604. [Google Scholar] [CrossRef] [PubMed]
- Riddell, E.; Lenihan, D. The role of cardiac biomarkers in cardio-oncology. Curr. Probl. Cancer 2018, 42, 375–385. [Google Scholar] [CrossRef]
- Iddagoda, M.T. The role of high-sensitive troponin measurement as a biomarker during the postoperative period for the detection of myocardial injury after non-cardiac surgery. J. Perioper. Pract. 2020. [Google Scholar] [CrossRef]
- Regan, B.; O’Kennedy, R.; Collins, D. Point-of-care compatibility of ultra-sensitive detection techniques for the cardiac biomarker troponin I—challenges and potential value. Biosensors 2018, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, R.P.; Neves, A.L.; Guimarães, H. Cardiac biomarkers in neonatology: BNP/NTproBNP, troponin I/T, CK-MB and myoglobin—A systematic review. J. Pediatr. Neonatal Individ. Med. 2017, 6, e060219. [Google Scholar] [CrossRef]
- Pyati, A.K.; Devaranavadagi, B.B.; Sajjannar, S.L.; Nikam, S.V.; Shannawaz, M.; Sudharani. Heart-type fatty acid binding protein: A better cardiac biomarker than CK-MB and myoglobin in the early diagnosis of acute myocardial infarction. J. Clin. Diagn. Res. 2015, 9, BC08–BC11. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, F.; Yang, X.; Wang, K.; Wang, H.; Deng, X. Sensitive point-of-care monitoring of cardiac biomarker myoglobin using aptamer and ubiquitous personal glucose meter. Biosens. Bioelectron. 2015, 64, 161–164. [Google Scholar] [CrossRef]
- Goetze, J.P.; Bruneau, B.G.; Ramos, H.R.; Ogawa, T.; de Bold, M.K.; de Bold, A.J. Cardiac natriuretic peptides. Nat. Rev. Cardiol. 2020, 17, 698–717. [Google Scholar] [CrossRef]
- Seliger, S. The Cardiorenal Syndrome: Mechanistic Insights and Prognostication with Soluble Biomarkers. Curr. Cardiol. Rep. 2020, 22, 114. [Google Scholar] [CrossRef] [PubMed]
- Pregenzer-Wenzler, A.; Abraham, J.; Barrell, K.; Kovacsovics, T.; Nativi-Nicolau, J. Utility of Biomarkers in Cardiac Amyloidosis. JACC Heart Fail. 2020, 8, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Gu, H.; Jeon, W.; Youn, H.; Her, J.; Kim, S.K.; Lee, J.; Shin, J.H.; Ban, C. Electrochemical Aptasensor of Cardiac Troponin i for the Early Diagnosis of Acute Myocardial Infarction. Anal. Chem. 2015, 87, 9869–9875. [Google Scholar] [CrossRef] [PubMed]
- Lopa, N.S.; Rahman, M.M.; Ahmed, F.; Ryu, T.; Sutradhar, S.C.; Lei, J.; Kim, J.; Kim, D.H.; Lee, Y.H.; Kim, W. Simple, low-cost, sensitive and label-free aptasensor for the detection of cardiac troponin I based on a gold nanoparticles modified titanium foil. Biosens. Bioelectron. 2019, 126, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Negahdary, M.; Behjati-Ardakani, M.; Heli, H. An electrochemical troponin T aptasensor based on the use of a macroporous gold nanostructure. Microchim. Acta 2019, 186, 377. [Google Scholar] [CrossRef]
- Negahdary, M.; Behjati-Ardakani, M.; Sattarahmady, N.; Yadegari, H.; Heli, H. Electrochemical aptasensing of human cardiac troponin I based on an array of gold nanodumbbells-Applied to early detection of myocardial infarction. Sens. Actuators B Chem. 2017, 252, 62–71. [Google Scholar] [CrossRef]
- Jo, H.; Her, J.; Lee, H.; Shim, Y.B.; Ban, C. Highly sensitive amperometric detection of cardiac troponin I using sandwich aptamers and screen-printed carbon electrodes. Talanta 2017, 165, 442–448. [Google Scholar] [CrossRef]
- Chekin, F.; Vasilescu, A.; Jijie, R.; Singh, S.K.; Kurungot, S.; Iancu, M.; Badea, G.; Boukherroub, R.; Szunerits, S. Sensitive electrochemical detection of cardiac troponin I in serum and saliva by nitrogen-doped porous reduced graphene oxide electrode. Sens. Actuators B Chem. 2018, 262, 180–187. [Google Scholar] [CrossRef]
- Qiao, X.; Li, K.; Xu, J.; Cheng, N.; Sheng, Q.; Cao, W.; Yue, T.; Zheng, J. Novel electrochemical sensing platform for ultrasensitive detection of cardiac troponin I based on aptamer-MoS2 nanoconjugates. Biosens. Bioelectron. 2018, 113, 142–147. [Google Scholar] [CrossRef]
- Luo, Z.; Sun, D.; Tong, Y.; Zhong, Y.; Chen, Z. DNA nanotetrahedron linked dual-aptamer based voltammetric aptasensor for cardiac troponin I using a magnetic metal-organic framework as a label. Microchim. Acta 2019, 186, 374. [Google Scholar] [CrossRef]
- Lang, M.; Luo, D.; Yang, G.; Mei, Q.; Feng, G.; Yang, Y.; Liu, Z.; Chen, Q.; Wu, L. An ultrasensitive electrochemical sensing platform for the detection of cTnI based on aptamer recognition and signal amplification assisted by TdT. RSC Adv. 2020, 10, 36396–36403. [Google Scholar] [CrossRef]
- Kumar, V.; Shorie, M.; Ganguli, A.K.; Sabherwal, P. Graphene-CNT nanohybrid aptasensor for label free detection of cardiac biomarker myoglobin. Biosens. Bioelectron. 2015, 72, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Emrani, A.S.; Abnous, K. A novel electrochemical aptasensor based on Y-shape structure of dual-aptamer-complementary strand conjugate for ultrasensitive detection of myoglobin. Biosens. Bioelectron. 2016, 80, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Brent, J.R.; Shorie, M.; Kaur, H.; Chadha, G.; Thomas, A.G.; Lewis, E.A.; Rooney, A.P.; Nguyen, L.; Zhong, X.L.; et al. Nanostructured aptamer-functionalized black phosphorus sensing platform for label-free detection of myoglobin, a cardiovascular disease biomarker. ACS Appl. Mater. Interfaces 2016, 8, 22860–22868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, Z.; Wang, L.; Guo, Y. Electrochemical aptasensor for myoglobin-specific recognition based on porphyrin functionalized graphene-conjugated gold nanocomposites. Sensors 2016, 16, 1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Xia, J.; Liu, H.; Zhang, F.; Wang, Z.; Lu, L. A new dual-signalling electrochemical aptasensor with the integration of “signal on/off” and “labeling/label-free” strategies. Sens. Actuators B Chem. 2017, 239, 166–171. [Google Scholar] [CrossRef]
- Nia, N.G.; Azadbakht, A. Nanostructured aptamer-based sensing platform for highly sensitive recognition of myoglobin. Microchim. Acta 2018, 185, 333. [Google Scholar] [CrossRef]
- Adeel, M.; Rahman, M.M.; Lee, J.-J. Label-free aptasensor for the detection of cardiac biomarker myoglobin based on gold nanoparticles decorated boron nitride nanosheets. Biosens. Bioelectron. 2019, 126, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, J.; Kang, M.; Huang, X.; Liu, Y.; Zhou, N.; Zhang, Z. Nanodiamonds and hydrogen-substituted graphdiyne heteronanostructure for the sensitive impedimetric aptasensing of myocardial infarction and cardiac troponin I. Anal. Chim. Acta 2021, 1141, 110–119. [Google Scholar] [CrossRef]
- Tao, D.; Shui, B.; Gu, Y.; Cheng, J.; Zhang, W.; Jaffrezic-Renault, N.; Song, S.; Guo, Z. Development of a label-free electrochemical aptasensor for the detection of Tau381 and its preliminary application in AD and non-AD patients’ sera. Biosensors 2019, 9, 84. [Google Scholar] [CrossRef] [Green Version]
- Shui, B.; Tao, D.; Cheng, J.; Mei, Y.; Jaffrezic-Renault, N.; Guo, Z. A novel electrochemical aptamer-antibody sandwich assay for the detection of tau-381 in human serum. Analyst 2018, 143, 3549–3554. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, H.; Liu, L.; Li, C.; Chang, Z.; Zhu, X.; Ye, B.; Xu, M. Fabrication of an antibody-aptamer sandwich assay for electrochemical evaluation of levels of β-amyloid oligomers. Sci. Rep. 2016, 6, 35186. [Google Scholar] [CrossRef] [PubMed]
- Negahdary, M.; Heli, H. An ultrasensitive electrochemical aptasensor for early diagnosis of Alzheimer’s disease, using a fern leaves-like gold nanostructure. Talanta 2019, 198, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Figueroa-Miranda, G.; Lyu, Z.; Zafiu, C.; Willbold, D.; Offenhäusser, A.; Mayer, D. Monitoring amyloid-Β proteins aggregation based on label-free aptasensor. Sens. Actuators B Chem. 2019, 288, 535–542. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, C.; Li, X.; Zhu, X.; Ye, B.; Xu, M. A sensitive aptasensor for the detection of β-amyloid oligomers based on metal-organic frameworks as electrochemical signal probes. Anal. Methods 2018, 10, 4430–4437. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Nameghi, M.A.; Ramezani, M.; Alibolandi, M.; Hassanzadeh-Khayat, M.; Emrani, A.S.; Abnous, K. A novel electrochemical aptasensor based on nontarget-induced high accumulation of methylene blue on the surface of electrode for sensing of α-synuclein oligomer. Biosens. Bioelectron. 2019, 123, 14–18. [Google Scholar] [CrossRef]
- Li, H.Y.; Jia, W.N.; Li, X.Y.; Zhang, L.; Liu, C.; Wu, J. Advances in Detection of Infectious Agents by Aptamer-based Technologies. Emerg. Microbes Infect. 2020, 9, 1671–1681. [Google Scholar] [CrossRef]
- Andryukov, B.G.; Besednova, N.N.; Romashko, R.V.; Zaporozhets, T.S.; Efimov, T.A. Label-free biosensors for laboratory-based diagnostics of infections: Current achievements and new trends. Biosensors 2020, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tuleouva, N.; Ramanculov, E.; Revzin, A. Aptamer-based electrochemical biosensor for interferon gamma detection. Anal. Chem. 2010, 82, 8131–8136. [Google Scholar] [CrossRef] [Green Version]
- Crulhas, B.P.; Hadley, D.; Liu, Y.; Shin, D.-S.; Stybayeva, G.; Imanbekova, M.; Hill, A.E.; Pedrosa, V.; Revzin, A. An electrochemical aptasensor for detection of bovine interferon gamma. Anal. Methods 2017, 9, 4527–4532. [Google Scholar] [CrossRef]
- Farid, S.; Meshik, X.; Choi, M.; Mukherjee, S.; Lan, Y.; Parikh, D.; Poduri, S.; Baterdene, U.; Huang, C.E.; Wang, Y.Y.; et al. Detection of Interferon gamma using graphene and aptamer based FET-like electrochemical biosensor. Biosens. Bioelectron. 2015, 71, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rahimian, A.; Krylyuk, S.; Vu, T.; Crulhas, B.; Stybayeva, G.; Imanbekova, M.; Shin, D.S.; Davydov, A.; Revzin, A. Nanowire Aptasensors for Electrochemical Detection of Cell-Secreted Cytokines. ACS Sens. 2017, 2, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kwa, T.; Revzin, A. Simultaneous detection of cell-secreted TNF-α and IFN-γ using micropatterned aptamer-modified electrodes. Biomaterials 2012, 33, 7347–7355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Liu, F.; Zhang, L.; Liu, L.; Liu, C.; Pu, X. An electrochemical strategy with molecular beacon and hemin/G-quadruplex for the detection of Clostridium perfringens DNA on screen-printed electrodes. RSC Adv. 2014, 4, 57064–57070. [Google Scholar] [CrossRef]
- Rashid, S.; Nawaz, M.H.; Marty, J.L.; Hayat, A. Label free ultrasensitive detection of NS1 based on electrochemical aptasensor using polyethyleneimine aggregated AuNPs. Microchem. J. 2020, 158, 105285. [Google Scholar] [CrossRef]
- Xu, A.; Wang, Z.; Zhang, C. Advance in development of methods for sensitive detection of tumor-related biomarkers. Kexue Tongbao Chin. Sci. Bull. 2017, 62, 859–870. [Google Scholar] [CrossRef]
- Safarpour, H.; Dehghani, S.; Nosrati, R.; Zebardast, N.; Alibolandi, M.; Mokhtarzadeh, A.; Ramezani, M. Optical and electrochemical-based nano-aptasensing approaches for the detection of circulating tumor cells (CTCs). Biosens. Bioelectron. 2020, 148, 111833. [Google Scholar] [CrossRef]
- Qu, L.; Xu, J.; Tan, X.; Liu, Z.; Xu, L.; Peng, R. Dual-Aptamer Modification Generates a Unique Interface for Highly Sensitive and Specific Electrochemical Detection of Tumor Cells. ACS Appl. Mater. Interfaces 2014, 6, 7309–7315. [Google Scholar] [CrossRef]
- Shen, H.; Yang, J.; Chen, Z.; Chen, X.; Wang, L.; Hu, J.; Ji, F.; Xie, G.; Feng, W. A novel label-free and reusable electrochemical cytosensor for highly sensitive detection and specific collection of CTCs. Biosens. Bioelectron. 2016, 81, 495–502. [Google Scholar] [CrossRef]
- Cao, J.; Zhao, X.-P.; Younis, M.R.; Li, Z.-Q.; Xia, X.-H.; Wang, C. Ultrasensitive Capture, Detection, and Release of Circulating Tumor Cells Using a Nanochannel–Ion Channel Hybrid Coupled with Electrochemical Detection Technique. Anal. Chem. 2017, 89, 10957–10964. [Google Scholar] [CrossRef]
- Zhai, T.-T.; Ye, D.; Zhang, Q.-W.; Wu, Z.-Q.; Xia, X.-H. Highly Efficient Capture and Electrochemical Release of Circulating Tumor Cells by Using Aptamers Modified Gold Nanowire Arrays. ACS Appl. Mater. Interfaces 2017, 9, 34706–34714. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Tang, X.; Wang, Y.; Fu, W.; Chang, K.; Chen, M. Target-triggered “signal-off” electrochemical aptasensor assisted by Au nanoparticle–modified sensing platform for high-sensitivity determination of circulating tumor cells. Anal. Bioanal. Chem. 2020, 412, 8107–8115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, H.; Yang, M.; Liao, L. Electrochemical assay for detection of circulating tumor cells based on LiFePO4 as electrochemical probe. Mater. Lett. 2020, 276, 128219. [Google Scholar] [CrossRef]
- Shen, H.; Deng, W.; He, Y.; Li, X.; Song, J.; Liu, R.; Liu, H.; Yang, G.; Li, L. Ultrasensitive aptasensor for isolation and detection of circulating tumor cells based on CeO2@Ir nanorods and DNA walker. Biosens. Bioelectron. 2020, 168, 112516. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Wu, D.; Yu, H.; Sun, W.; Yi, X.; Liu, L. Magnetic bead-based electrochemical and colorimetric assays of circulating tumor cells with boronic acid derivatives as the recognition elements and signal probes. Talanta 2021, 221, 121640. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.B. Tumor markers. Physiology, pathobiology, technology, and clinical applications. Clin. Chim. Acta 2003, 330, 185–186. [Google Scholar] [CrossRef]
- Shu, H.; Wen, W.; Xiong, H.; Zhang, X.; Wang, S. Novel electrochemical aptamer biosensor based on gold nanoparticles signal amplification for the detection of carcinoembryonic antigen. Electrochem. Commun. 2013, 37, 15–19. [Google Scholar] [CrossRef]
- Xue, S.; Yi, H.; Jing, P.; Xu, W. Dendritic Pt@Au nanowires as nanocarriers and signal enhancers for sensitive electrochemical detection of carcinoembryonic antigen. RSC Adv. 2015, 5, 77454–77459. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Guo, Y.; Dong, C. Label-free Electrochemical Aptasensor for Carcino-embryonic Antigen Based on Ternary Nanocomposite of Gold Nanoparticles, Hemin and Graphene. Electroanalysis 2016, 28, 1023–1028. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Xia, Y.; Li, J.; Wang, E. Electrochromic sensing platform based on steric hindrance effects for CEA detection. Analyst 2016, 141, 3985–3988. [Google Scholar] [CrossRef]
- Wen, W.; Huang, J.-Y.; Bao, T.; Zhou, J.; Xia, H.-X.; Zhang, X.-H.; Wang, S.-F.; Zhao, Y.-D. Increased electrocatalyzed performance through hairpin oligonucleotide aptamer-functionalized gold nanorods labels and graphene-streptavidin nanomatrix: Highly selective and sensitive electrochemical biosensor of carcinoembryonic antigen. Biosens. Bioelectron. 2016, 83, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xue, S.; Jing, P.; Xu, W. A sensitive impedimetric platform biosensing protein: Insoluble precipitates based on the biocatalysis of manganese(III) meso-tetrakis (4-N-methylpyridiniumyl)-porphyrinin in HCR-assisted dsDNA. Biosens. Bioelectron. 2016, 86, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wan, Y.; Deng, S.; Yang, S.; Su, Y.; Fan, C.; Aldalbahi, A.; Zuo, X. Aptamer-initiated on-particle template-independent enzymatic polymerization (aptamer-OTEP) for electrochemical analysis of tumor biomarkers. Biosens. Bioelectron. 2016, 86, 536–541. [Google Scholar] [CrossRef] [PubMed]
- He, B. Differential pulse voltammetric assay for the carcinoembryonic antigen using a glassy carbon electrode modified with layered molybdenum selenide, graphene, and gold nanoparticles. Microchim. Acta 2017, 184, 229–235. [Google Scholar] [CrossRef]
- Shekari, Z.; Zare, H.R.; Falahati, A. Developing an impedimetric aptasensor for selective label–free detection of CEA as a cancer biomarker based on gold nanoparticles loaded in functionalized mesoporous silica films. J. Electrochem. Soc. 2017, 164, B739–B745. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Duan, F.-H.; Tian, J.-Y.; He, J.-Y.; Yang, L.-Y.; Zhao, H.; Zhang, S.; Liu, C.-S.; He, L.-H.; Chen, M.; et al. Aptamer-Embedded Zirconium-Based Metal–Organic Framework Composites Prepared by De Novo Bio-Inspired Approach with Enhanced Biosensing for Detecting Trace Analytes. ACS Sens. 2017, 2, 982–989. [Google Scholar] [CrossRef]
- Si, Z.; Xie, B.; Chen, Z.; Tang, C.; Li, T.; Yang, M. Electrochemical aptasensor for the cancer biomarker CEA based on aptamer induced current due to formation of molybdophosphate. Microchim. Acta 2017, 184, 3215–3221. [Google Scholar] [CrossRef]
- Xiang, J.; Pi, X.; Chen, X.; Xiang, L.; Yang, M.; Ren, H.; Shen, X.; Qi, N.; Deng, C. Integrated signal probe based aptasensor for dual-analyte detection. Biosens. Bioelectron. 2017, 96, 268–274. [Google Scholar] [CrossRef]
- Jiang, W.; Liu, L.; Zhang, L.; Guo, Q.; Cui, Y.; Yang, M. Sensitive immunosensing of the carcinoembryonic antigen utilizing aptamer-based in-situ formation of a redox-active heteropolyacid and rolling circle amplification. Microchim. Acta 2017, 184, 4757–4763. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, S.; Gao, J.; Zhao, J.; Xue, S.; Xu, W. Glucose oxidase-initiated cascade catalysis for sensitive impedimetric aptasensor based on metal-organic frameworks functionalized with Pt nanoparticles and hemin/G-quadruplex as mimicking peroxidases. Biosens. Bioelectron. 2017, 98, 83–90. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhao, L.; Lei, W.; Wen, W.; Wang, Y.J.; Bao, T.; Xiong, H.Y.; Zhang, X.H.; Wang, S.F. A high-sensitivity electrochemical aptasensor of carcinoembryonic antigen based on graphene quantum dots-ionic liquid-nafion nanomatrix and DNAzyme-assisted signal amplification strategy. Biosens. Bioelectron. 2018, 99, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, G.; Zou, L.; Lei, S.; Yu, Q.; Ye, B. Highly active DNAzyme-peptide hybrid structure coupled porous palladium for high-performance electrochemical aptasensing platform. Sens. Actuators B Chem. 2018, 259, 372–379. [Google Scholar] [CrossRef]
- Wang, Q.L.; Cui, H.F.; Song, X.; Fan, S.F.; Chen, L.L.; Li, M.M.; Li, Z.Y. A label-free and lectin-based sandwich aptasensor for detection of carcinoembryonic antigen. Sens. Actuators B Chem. 2018, 260, 48–54. [Google Scholar] [CrossRef]
- Soung, Y.; Ford, S.; Zhang, V.; Chung, J. Exosomes in Cancer Diagnostics. Cancers 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Sun, R.; He, P.; Zhang, X. Ultrasensitive Detection of Exosomes by Target-Triggered Three-Dimensional DNA Walking Machine and Exonuclease III-Assisted Electrochemical Ratiometric Biosensing. Anal. Chem. 2019, 91, 14773–14779. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, L.; Wan, S.; Cansiz, S.; Cui, C.; Liu, Y.; Cai, R.; Hong, C.; Teng, I.T.; Shi, M.; et al. Aptasensor with Expanded Nucleotide Using DNA Nanotetrahedra for Electrochemical Detection of Cancerous Exosomes. ACS Nano 2017, 11, 3943–3949. [Google Scholar] [CrossRef]
- Zhou, Q.; Rahimian, A.; Son, K.; Shin, D.-S.; Patel, T.; Revzin, A. Development of an aptasensor for electrochemical detection of exosomes. Methods 2016, 97, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Chen, H.; Jiang, J.; Zhang, H.; Cai, C.; Shen, Q. Highly Sensitive Electrochemical Detection of Tumor Exosomes Based on Aptamer Recognition-Induced Multi-DNA Release and Cyclic Enzymatic Amplification. Anal. Chem. 2018, 90, 4507–4513. [Google Scholar] [CrossRef]
- An, Y.; Jin, T.; Zhu, Y.; Zhang, F.; He, P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens. Bioelectron. 2019, 142, 111503. [Google Scholar] [CrossRef]
- Yin, X.; Hou, T.; Huang, B.; Yang, L.; Li, F. Aptamer recognition-trigged label-free homogeneous electrochemical strategy for an ultrasensitive cancer-derived exosome assay. Chem. Commun. 2019, 55, 13705–13708. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Han, B.; Wang, Y.; Dai, Y.; Zhao, J. A catalytic molecule machine-driven biosensing method for amplified electrochemical detection of exosomes. Biosens. Bioelectron. 2019, 141, 111397. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liao, C.; Zuo, P.; Liu, Z.; Ye, B.-C. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.; Eck, S.L. EpCAM: A New Therapeutic Target for an Old Cancer Antigen. Cancer Biol. Ther. 2003, 2, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yang, B.; Qian, K.; Qiao, L.; Liu, Y.; Liu, B. Sensitive electrochemical aptasensor for detecting EpCAM with silica nanoparticles and quantum dots for signal amplification. J. Electroanal. Chem. 2020, 856, 113655. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, W.; Shang, B.; Wei, J.; Chen, L.; Guo, X.; Ran, F.; Chen, W.; Ding, X.; Xu, Y.; et al. Ultrasensitive amperometric aptasensor for the epithelial cell adhesion molecule by using target-driven toehold-mediated DNA recycling amplification. Microchim. Acta 2018, 185, 202. [Google Scholar] [CrossRef]
- Cao, H.; Ye, D.; Zhao, Q.; Luo, J.; Zhang, S.; Kong, J. A novel aptasensor based on MUC-1 conjugated CNSs for ultrasensitive detection of tumor cells. Analyst 2014, 139, 4917. [Google Scholar] [CrossRef]
- Ma, F.; Ho, C.; Cheng, A.K.H.; Yu, H.-Z. Immobilization of redox-labeled hairpin DNA aptamers on gold: Electrochemical quantitation of epithelial tumor marker mucin 1. Electrochim. Acta 2013, 110, 139–145. [Google Scholar] [CrossRef]
- Zheng, T.; Tan, T.; Zhang, Q.; Fu, J.-J.; Wu, J.-J.; Zhang, K.; Zhu, J.-J.; Wang, H. Multiplex acute leukemia cytosensing using multifunctional hybrid electrochemical nanoprobes at a hierarchically nanoarchitectured electrode interface. Nanoscale 2013, 5, 10360. [Google Scholar] [CrossRef]

| Cardiac Biomarker | Sensor Specification | Method | Sensing Strategy | Sample Matrices | Detection Rang | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|---|
| Mb | polystyrene microplate | personal glucose meter (PGM) readout | antibody–Mb–aptamer sandwich | serum samples | 50 pM–200 nM | 50 pM | [71] |
| cTnI | Au electrode/thiolated apt. | SWV, Fc-SiNPs | label-free | buffer | 1−10,000 pM | 1.0 pM | [75] |
| cTnI | AuNPs, Ti Foil/HS-apt. | DPV, [Fe(CN)6]3−/4− | Label-free | human serum samples | 1–1100 pM | 0.18 pM | [76] |
| cTnI | Au electrode/macroporous Au nanostructure/Apt. | DPV, [Fe(CN)6]3−/4− | label-free | spiked human serum samples | 0.05–5.0 ng mL−1 | 23 pg mL−1 | [77] |
| TnI | array of Au nanodumbbells/76-mer TnI aptamer | DPV [Fe(CN)6]3−/4− | label-free | blood serum samples | 0.05–500 ng mL−1 | 8.0 pg mL−1 | [78] |
| cTnI | SPCE/AuNPs/electropolymerized conductive polymer/Apt. | amperometry, hydrazine, H2O2 | apt. pair-based sandwich type | serum | 1–100 pM (0.024–2.4 ng/mL) | 1.0 pM (24 pg/mL) | [79] |
| cTnI | GC/N-prGO/COOH/PEG-apt. | DPV | Label-free | serum, saliva | 0.001–100 ng mL−1 | 0.88 pg mL−1 | [80] |
| cTnI | Apt.–MoS2 nanosheets | EIS, [Fe(CN)6]3−/4− | label-free | human blood samples | 10 pM to 1.0 μM | 0.95 pM | [81] |
| Au@SiO2@Au/aptamer- | 10 pM–10.0 μM | 1.23 pM | |||||
| cTnI | DNA nanotetrahedron (NTH)/dual-apt. (dAPT), magnetic metal organic frameworks (mMOFs), Fe3O4@UiO-66, Au@Pt NPs | DPV hydroquinone, H2O2 | sandwich-structure | human serum | 0.01–100 ng·mL−1 | 5.7 pg mL−1 | [82] |
| cTnI | Au electrode/apt. probe 2 (P2) | SWV MB | sandwich-structure | human serum, urine saliva | 0.5–100 ngmL−1 | 0.04 ngmL−1 | [83] |
| Mb | apt./rGO/CNT nanohybrid electrodes | CV direct electron transfer Mb | label-free | buffer | 1 ng/mL–4 µg/mL | ~0.34 ng/mL | [84] |
| Mb | Y-shape structure of dual-aptamer (DApt)- | DPV [Fe(CN)6]3−/4− | label-free | human blood serum | 100 pM–40 nM | 27 pM | [85] |
| Mb | apt.–black phosphorus nanostructured electrode | CV | direct electron transfer Mb | serum samples | 1 pg mL−1–16 µg mL−1 | 0.524 pg mL−1 | [86] |
| Mb | meso-tetra (4-carboxyphenyl) porphyrin-AuNPs(TCPP–Gr/AuNPs)/apt. Mb | DPV | electron transfer by Mb | human serum samples | 2.0 × 10−11–7.7 × 10−7 M | 6.7 × 10−12 M | [87] |
| Lys | Au electrode/Lys apt. | SWV Fc, [Ru(NH3)6]3+ | signal on/off, label-free | human serum | 1.0 × 10−11–1.0 × 10−7 M | 0.8 pM | [88] |
| Mb | GCE, PtSnNP CNTs/Mb-apt. | Amperometry, [Fe(CN)6]3−/4- | conformational change | Spiked human serum samples | 0.01–1 and 10 nM–200 nM | 2.2 ± 0.1 pM | [89] |
| Mb | Apt/AuNPs/BNNSs/FTO | DPV [Fe(CN)6]3−/4− | label free | human serum Hb, GOx, Ins., SOx | 0.1–100 µg/mL | 34.6 ng/mL | [90] |
| cTnI Mb | HsGDY@NDs/apt.strand | EIS [Fe(CN)6]3−/4− | label-free | human serum | cTnI 0.00001–100 ng mL−1 Mb 0.01–1000 pg mL−1 | cTnI 9.04 fg mL−1 Mb 9.04 fg mL−1 | [91] |
| AD Biomarkers | Sensor Specification | Method | Sensing Strategy | Sample Matrices | Detection Rang | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|---|
| tau 381 | apt./carboxyl Gr/th./AuNPs GCE | DPV Th. redox probe | label-free | 10 patients’ serum samples | 1.0–100 pM | 0.70 pM | [92] |
| tau 381 | tau antibody (anti-tau)/apt.specific to tau-381 | EIS, CV, DPV AuNPs signal amplification | Apt.-antibody sandwich | AD patients serum | 0.5–100 pM | 0.42 pM | [93] |
| AβO | β oligomers antibodies/nanocomposite AuNPs/apt. | DPV Th. redox probe | antibody-apt. sandwich | artificial cerebrospinal fluid (CSF) | 0.5–30 nM | 100 pM | [94] |
| AβO | SAM ssDNA apt./Au electrode | EIS [Fe(CN)6]3−/4− | label-free | artificial CSF | 0.1–500 nM | 0.03 nM | [96] |
| AβO | AuNFs/GCE/HS-apt. | DPV MOFs signalprobe | sandwich | artificial CSF | 1 nM–2 µM | 0.45 nM | [97] |
| α-synuclein | Apt./SAM | DPV MB | label-free | human serum samples | 60 pM–150 nM | 10 pM | [98] |
| AβO ATP | nanostructured multielectrode arrays (MEAs); dual-apt. | Amperometry Fc | signal-on/signal-off | artificial CSF | 1 pM–200 nM 0.01 nM–1000 nM | 0.3 pM 0.002 nM | [99] |
| Infectious Disease Biomarkers | Sensor Specification | Method | Sensing Strategy | Sample Matrices | Detection Range | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|---|
| IFN-γ | SAM DNA hairpin IFN-γ-apt./MB/Au electrode | SWV | conformational changes | serum proteins | 0.06–10 nM | 0.06 nM | [101] |
| bovine interferon gamma (BoIFN-γ) | SAM IFN-γ-binding apt. MB | SWV | conformational change | pristine buffer, blood | 1–230 ng/mL | pristine buffer 0.1 nM blood 0.9 nM | [102] |
| IFN-γ | IFN-γ apt./Gr surface | conductance | change in charge distribution in electrolyte | BSA, papain | 0 nM–5 μM | 83 pM | [103] |
| IFN-γ | SiNWs/Au/thiolated apt. | SWV MB | conformational change, Switch off | blood serum | nanowire 3 pg/mL for 5000 cells—planar 90 pg/mL for 30,000 cells | 0.14 ng/mL, 0.8 ng/mL | [104] |
| IFN-γ TNF- α | micropatterned Au electrodes arrays/cytokine-binding apt./microfluidic device | SWV | conformational change | immune cells | 0.0–100 ng/mL (5.8 nM) | 10 ng/mL (0.58 nM) | [105] |
| C. perfringens pathogen | capture probe, streptavidin (SA), hemin/G-quadruplex/Fe3O4 nanocomposites | DPV | sandwich | Real biological samples | 10−12–10−6 M | 10−12 M | [106] |
| NS-1 protein | Pencil electrode (PE)/(BE)/PEI/AuNPs/apt. | CV [Fe (CN)6]−3/−4 | label-free, structural switching | spiked human serum sample | 3–160 ng mL−1 | 0.3 ng mL−1 | [107] |
| CTCs | Sensor Specification | Method | Sensing Strategy | Sample Matrices/ Interferents | Detection Range | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|---|
| MEAR cancer cells | dual-apt. TLS1c, TLS11/amide bond/GCE | DPV [Fe(CN)6]3−/4− | label-free | blood cells | 1−14 MEAR cells | single MEAR cell/109 whole blood cells | [110] |
| CTCs | EpCAM apt./MCH/Au electrode | EIS [Fe(CN)6]3−/4− | label-free | real blood sample | 30–1 × 106 cells L−1 | 10 cells mL−1 | [111] |
| acute leukemia CCRF-CEM | Apt./array nanochannel–ionchannel hybrid | LSV | label-free | k562 cells, Ramos cells | 1 × 102–2 × 106 cells mL−1 | 100 cells mL−1 | [112] |
| MCF-7 circulating tumor cells | MUC-1 apt./rGO/AuNPs composites | DPV CuO NPs, H2O2 | label-free | serum sample | 50–7 × 103 cells mL−1 | 27 cells mL−1 | [64] |
| Ramos, CCRF-CEM cells | Apt.–AuNPs Array/Magnetic Gr Nanosheets | SWASV multiplexed detection | label-free | human whole blood | 5–500 cells mL−1 | Ramos 4 cells mL−1 CCRF-CEM cells 3 cells mL−1 | [62] |
| CTCs K562 cell | AuNPs/Au electrode, MB–apt. | Alternating current voltammetry (ACV) MB | signal-off | U937, Jurkat cells | 1 × 102–1 × 106 cells mL−1 | 23 cells mL−1 | [114] |
| Breast cancer cell MCF-7 | Fe3O4 magnetic nanospheres (MNs)/EpCAM antibody | SWV molybdophosphate (PMo12O40) redox label | antibody–aptamer sandwich type | whole blood samples | 3–10,000 cell mL−1 | 1 cell mL−1 | [115] |
| MCF-7 cells | Membrane protein MUC1-targeting apt. | dual signal, (Ce@IrNRs) enzyme-free DNA walker | enzyme-free DNA walker | whole blood samples | 2–2 × 106 cells mL−1 | 1 cell mL−1 | [116] |
| CTCs MCF-7 | Apt. magnetic beads MBs | DPV ferroceneboronic acid (FcBA) | label-free strategy | blood sample | 50−2 × 104 cells | 50 cells | [117] |
| Sensor Specification | Method | Sensing Strategy | Sample Matrices | Detection Range | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|
| apt1/Au | DPV Fc | sandwich-type | human serum spiked | 1–200 ng/mL | 0.5 ng/mL | [119] |
| dendritic Pt@AuNWs nanocarriers/thiol- CEA apt. 2 | DPV Tb, Pt@AuNWs H2O2 | sandwich-type | human serum sample | 0.001–80 ng mL−1 | 0.31 pg mL−1 | [120] |
| Au/DNA1/MCH/DNA2/DNA3-MB electrode | SWV MB | label-free | tumor necrosis factor-α (TNF-α), thrombin | 10–100 nM (MUC1) 30–300 ng mL−1 (CEA) | 0.13 nM (MUC1) 2.75 ng mL−1 (CEA) | [121] |
| aptamer/hemin and graphene nanosheets (AuNPs-HGNs)/GCE | DPV electron transfer of hemin | label-free | human serum albumin HSA, thrombin (TB), lysozyme (LYS), insulin (INS) | 0.0001–10 ng mL−1 | 40 fg mL−1 | [121] |
| MCH/ITO/Au film CEA apt./anode pole bipolar electrode (BPE) | EIS PB | structural change | human serum; AFP, BSA, thrombin | 1–10 ng mL−1 | - | [122] |
| hairpin oligonucleotide apt./Au nanorods/Gr-streptavidin nanomatrix/GCE | DPV HRP, H2O2, o-Phenylenediamine | switch-on | 8 human serum samples, lung diseases patients | 5 pg mL−1–50 ng mL−1 | 1.5 pg mL−1 | [123] |
| PtPdNWs/CEA apt. | EIS oxidation DAB | label-free | human serum samples | 0.1–0.5 ng mL−1 and 1–40 ng mL−1 | 0.030 pg mL−1 | [124] |
| BSA/aptamer/AuNPs /MoSe2-Gr/GCE | DPV [Fe(CN)6]3−/4− | structural change switch-on | HAS, IgE, AFP, Thrombin, LDL | 0.1 pg mL−1–100 ng mL−1 | 0.03 pg mL−1 | [125] |
| amine–Apt./AuNPs/Mesoporous Silica Films/GCE | EIS [Fe(CN)6]3−/4− | structural change | patient, healthy human serum samples | 1.0 × 10−3−100.0 ng mL−1 | 9.8 × 10−4 ng mL−1 | [126] |
| 509-MOFs@Apt. strand | EIS [Fe(CN)6]3-/4- | conformational changes, label-free | human serum sample | 0.001–0.50 ng mL−1 | CEA 0.40 pg mL−1, thrombin 0.37 pg mL−1, Kanamycin 0.21 pg mL−1 | [127] |
| GCE/Au nanorods/Antibody | redox-active PMo12O40 | sandwich-type | human serum samples | 0.1 pg mL−1–10 ng mL−1 | 0.05 pg mL−1 | [128] |
| MCH/apt./(ISP)/AuNPs/GCE | SWV MB Fc | DNA structural switching | HAS, HIgG | 1–1 μg mL−1 (CEA), 5 nM–1 μM (MUC1) | 0.517 ng mL−1 (CEA), 1.06 nM (MUC1) | [129] |
| Zr-MOF AgNCs@Apt@UiO-66, AgNCs@CEA-apt. | EIS, DPV, SPR | label-free | human serum samples | 0.01−10 ng mL−1 (EIS, DPV) 1.0−250 ng mL−1 (SPR) | 8.88 (EIS) 4.93 (DPV) pg mL−1 0.3 ng mL−1 (SPR) | [130] |
| GCE/AuNP–apt. | SWV redox-active PMo12O40 rolling circle amplification (RCA) | sandwich-type | human serum samples | 0.5 pg mL−1–1 ng mL−1 | 0.1 pg mL−1 | [131] |
| GCE/apt./CuMOF/Pt | EIS 3DAB, insoluble precipitates (IPs) | sandwich-type | human serum samples | 0.05 pg mL−1–20 ng mL−1 | 0.023 pg mL−1 | [132] |
| GQDs-IL-NF/glassy carbon electrode/single-stranded DNA | DPV MB, Pb2+ DNAzyme- signal amplification | label- free | serum samples; CEA, PSA, BSA, MUC1 | 0.5 fg mL−1–0.5 ng mL−1 | 0.34 fg mL−1 | [133] |
| ADH/hemin/G-quadruplex (hGQ-peptide)/apt./PdNPs/GCE | DPV Tb, ADH | sandwich-type | serum sample; AFP, BSA, Hb | 0.0001–100 ng mL−1 | 20 fg mL−1 | [134] |
| MCH–Apt./Au | DPV HRP label | label-free, sandwich-type | human blood serum samples; BSA, HSA, γ-glubulin, AFP, CRP | 5–40 ng mL−1 | 3.4 ng mL−1 | [135] |
| Exosomes | Sensor Specification | Method | Sensing Strategy | Sample Matrices | Detection Range | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|---|
| MCF-7 exosomes | Apt./2D Ti3C2 MXenes nanosheets | Electrogenerated chemiluminescence (ECL) | sandwich-type | serum | 5 × 102–5 × 106 particles μL−1 | 125 particles μL−1 | [137] |
| hepatocellular exosomes | Apt.-nanotetrahedron (NTH) SAM/Au electrode | SWV [Fe(CN)6]−3/−4 | sandwich-type | membrane proteins | 105–1012 exosomes/mL | 1 × 104/mL | [138] |
| exosomes | exosome transmembrane protein CD63 apt./Au electrode array, microfluidic system | SWV MB redox label | switch-off | fetal bovine serum | 1 × 108 particles/mL | 1 × 106 particles/mL | [139] |
| tumor exosomes LNCaP cells | DNAs/Au electrode | DPV Ru(NH3)63− signal reporter | “turn-off” signal | MCF-7 cell, HeLa cell-derived exosomes | 1000–120,000 particles/µL | 70 particles/µL | [140] |
| Exosomes | CD63 apt./GCE/Cu (I)-catalyzed click chemistry | DPV HRP, o-phenylene-diamine (OPD), H2O2 | label-free | human serum | 1.12 × 102 to 1.12 × 108 particles/μL | 96 particles/μL | [141] |
| cancer-exosome | ITO electrode | DPV DOX redox-active indicator | label-free “signal-on” strategy | biological, clinical samples | 3.4 × 104–3.4 × 108 particles per mL | 1.2 × 104 particles per mL | [142] |
| HepG2-exosomes | Au electrode/apt. | SWV [Fe(CN)6]−3/−4 | displacement reaction | serum samples | 1 × 105–5 × 107 particles/mL | 1.72 × 104 particles/mL | [143] |
| CD63 positive exosomes | ITO electrode microfluidic on-chip platform (ExoPCD-chip) | DPV [Fe(CN)6]−3/−4, th. enhanced cathodic current | label-free | human serum and liver cancer patients, healthy controls | 7.61 × 104 particles/mL to 7.61 × 108 particles/mL | 4.39 × 103 particles/mL | [144] |
| Circulating Tumor Cells Biomarkers | Sensor Specification | Method | Sensing Strategy | Sample Matrices/Tested Interferents | Detection Range | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|---|
| EpCAM | Au electrode/2-mercaptoethanol/apt. | stripping SWV CdSe NPs signal | sandwich-type | added CK-19, thrombin | 10 aM to 100 pM | 10 aM | [146] |
| EpCAM | EpCAM apt. Hairpin probe 1 (Hp1) 5′-SH/SAM/MCH/Au electrode | SWV MB | switch-on | spiked human serum, urine, saliva | 0.1–20 ng mL−1 | 20 pg mL−1 | [147] |
| human colon cancer DLD-1 cells | MUC-1 apt./CNSs | EIS, CV [Fe(CN)6]−3/−4 | label-free | human astrocyte (HA) cells, DLD-1 cells | 1.25 × 102–1.25 × 106 cells per mL | 40 cells per mL | [148] |
| epithelial tumor marker mucin 1 (MUC1) | MB-hairpin DNA apt./Au electrode | SWV | conformational change switch-off | added lysozyme, cytochrome c, and BSA | up to 1.5 M | 50 nM | [149] |
| human leukemia cell line (HL-60) lymphocytic leukemia cells (CEM cells) | dual apt./multilayered graphene–AuNPs electrode | DPV HRP redox tags | sandwich-type | Hela, K562, normal red blood cells | 5 × 102–1 × 107 cells per mL | 350 cells per mL | [150] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radi, A.-E.; Abd-Ellatief, M.R. Electrochemical Aptasensors: Current Status and Future Perspectives. Diagnostics 2021, 11, 104. https://doi.org/10.3390/diagnostics11010104
Radi A-E, Abd-Ellatief MR. Electrochemical Aptasensors: Current Status and Future Perspectives. Diagnostics. 2021; 11(1):104. https://doi.org/10.3390/diagnostics11010104
Chicago/Turabian StyleRadi, Abd-Elgawad, and Maha Ragaa Abd-Ellatief. 2021. "Electrochemical Aptasensors: Current Status and Future Perspectives" Diagnostics 11, no. 1: 104. https://doi.org/10.3390/diagnostics11010104






