Abstract
Accelerating antimicrobial susceptibility testing (AST) is a priority in the development of novel microbiological methods. The MALDI-TOF MS-based direct-on-target microdroplet growth assay (DOT-MGA) has recently been described as a rapid phenotypic AST method. In this proof-of-principle study, we expanded this method to simultaneously test 24 antimicrobials. An Enterobacterales panel was designed and evaluated using 24 clinical isolates. Either one or two (only for antimicrobials with the EUCAST “I” category) breakpoint concentrations were tested. Microdroplets containing bacterial suspensions with antimicrobials and growth controls were incubated directly on the spots of a disposable MALDI target inside a humidity chamber for 6, 8 or 18 h. Broth microdilution was used as the standard method. After 6 and 8 h of incubation, the testing was valid (i.e., growth control was successfully detected) for all isolates and the overall categorical agreement was 92.0% and 92.7%, respectively. Although the overall assay performance applying short incubation times is promising, the lower performance with some antimicrobials and when using the standard incubation time of 18 h indicates the need for thorough standardization of assay conditions. While using “homebrew” utensils and provisional evaluation algorithms here, technical solutions such as dedicated incubation chambers, tools for broth removal and improved software analyses are needed.
1. Introduction
Rapid microbiological diagnostics are of considerable importance, as they contribute to the optimization of patient management [1] and improve clinical outcome [2,3]. With the introduction of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) into the diagnostic routine, considerable acceleration of microbial identification has been achieved [4,5]. However, such progress is lacking in routine antimicrobial susceptibility testing (AST), which is at least as important for therapeutic decisions as identification [6]. The AST findings are usually only available on the next day after test initiation [7].
In recent years, several techniques have been attempted to determine microbial resistance by MALDI-TOF MS [8,9]. Indeed, combining identification and AST on a single platform could result in synergistic effects for workflow and cost saving [6]. However, the ultimate goal is to develop a phenotypic AST assay that is universal and independent from resistance mechanisms [10,11]. The AST results should be available within a working shift to enable prompt adjustment of antimicrobial treatment on the same day [6].
The MALDI-TOF MS-based direct-on-target microdroplet growth assay (DOT-MGA) has recently been suggested as a rapid universal phenotypic AST method [11,12,13,14]. Initially, Idelevich et al. [12] introduced the DOT-MGA to determine carbapenem susceptibility in Klebsiella pneumoniae and Pseudomonas aeruginosa while microdroplets of bacterial suspension with and without the addition of antibiotics were incubated on a surface of a MALDI target and, after a short incubation time, growth or no growth of bacteria were detected by MALDI-TOF MS measurement to determine susceptibility or resistance. Further applications followed for various antibiotics and bacterial species [11,12,13,14].
In this study, we expanded this method to the proof-of-principle simultaneous testing of multiple antibiotics at breakpoint concentrations. For this purpose, we designed and evaluated DOT-MGA antibiotic panels for Enterobacterales, a clinically very important bacterial group in which antibiotic resistance is increasing worldwide. The panel included the most clinically relevant antimicrobials with EUCAST breakpoints available for these organisms.
2. Materials and Methods
2.1. Bacterial Strains
The strain collection of Enterobacterales consisted of twelve prospectively and consecutively collected clinical isolates, as well as twelve consecutive clinical multidrug resistant (MDR) isolates. In total, 24 Enterobacterales isolates comprised Klebsiella pneumoniae (n = 8), Escherichia coli (n = 4), Enterobacter cloacae complex (n = 4), Klebsiella oxytoca (n = 2), Klebsiella aerogenes (n = 2), Citrobacter koseri (n = 2), Citrobacter freundii (n = 1) and Serratia marcescens (n = 1). All isolates originated from routine diagnostics of the Institute of Medical Microbiology, University Hospital Münster, Münster, Germany. Only one isolate per patient was included. Microorganisms were isolated from urine (n = 7), tracheal fluid (n = 3), superficial swab (n = 3), deep swab (n = 2), tissue (n = 2), implant (n = 2), feces (n = 2), sputum (n = 1), blood culture (n = 1) and bronchial lavage (n = 1).
2.2. Antimicrobials
Antibiotic panels were composed to include 24 antimicrobials (Table 1 and Table S1) for which EUCAST breakpoints (version 9.0) were available for Enterobacterales [15]. To allow complete categorization as susceptible, standard dosing regimen (S), susceptible, increased exposure (I) or resistant (R) according to EUCAST breakpoints [15], one concentration was tested for antimicrobials for which no I category exists, and two concentrations were tested for antimicrobials with the I category available from EUCAST [15]. For that, antibiotics contained in the respective wells of a Micronaut-S microtiter plate (MERLIN Diagnostika, Bornheim-Hersel, Germany) were used as described below.

Table 1.
Results of simultaneous DOT-MGA testing of multiple antibiotics vs. Enterobacterales, n = 24.
2.3. MALDI-TOF MS Direct-on-Target Microdroplet Growth Assay (DOT-MGA)
Bacterial suspensions with turbidity of 0.5 McFarland standard were prepared in 0.9% saline from cultures incubated overnight on Columbia blood agar. These suspensions were diluted 1:200 in cation-adjusted Mueller-Hinton broth (CAMHB, BD Diagnostic, Heidelberg, Germany) to produce inoculum of approximately 5 × 105 cfu/mL. Final inoculum size was confirmed by plating onto tryptic soy agar plates in triplicate and colony counting after overnight incubation (18–24 h). Several 100-µL samples of prepared inoculum were added into the wells of a Micronaut-S microtiter plate containing dried antibiotics (MERLIN Diagnostika, Bornheim-Hersel, Germany), followed by shaking the plate for 5 min at 300 rpm at room temperature to ensure complete dissolution of antibiotics. Several 6-µL samples of bacterial suspensions with antibiotics, as well as a growth control without antibiotic, were spotted in duplicate from the wells of a microtiter plate onto the spots of disposable MALDI targets (MBT Biotarget 96, Bruker Daltonics GmbH & Co. KG, Bremen, Germany) as microdroplets. For each isolate, three separate targets were prepared to test three different incubation times. The inoculated targets were incubated at 36°C for 6, 8 or 18 h in a plastic box (Bruker Daltonics GmbH & Co. KG, Bremen, Germany) containing 4 mL water to avoid evaporation of microdroplets. After incubation, medium was removed by absorptive pads, as previously described [12,16,17,18,19], but introducing the modification that the absorptive pad is applied directly from the top of microdroplets to remove broth from all the spots on a target in a single action (Figure 1). An amount of 1 μL MBT FAST Matrix (Bruker Daltonics GmbH & Co. KG, Bremen, Germany) containing an internal standard was added to each spot, and MALDI-TOF MS measurement performed twice for each spot with a MALDI Biotyper smart instrument (Bruker Daltonics GmbH & Co. KG, Bremen, Germany) and the DOT-MGA settings as previously described [19].
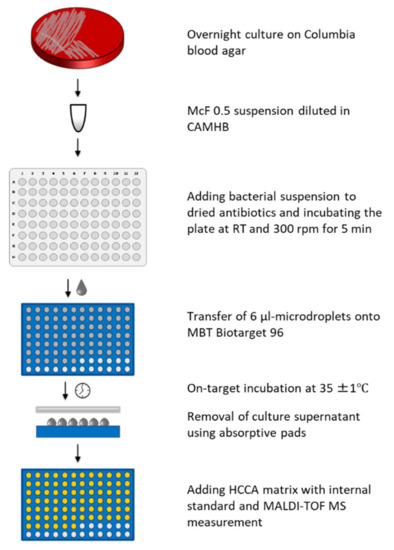
Figure 1.
Workflow of the MALDI-TOF MS direct-on-target microdroplet growth assay (DOT-MGA).
Mass spectra were interpreted using the MBT FAST prototype software (Bruker Daltonics GmbH & Co. KG, Bremen, Germany). In case of bacterial growth in a microdroplet with antibiotic, bacterial biomass is successfully detected by MALDI-TOF MS after the incubation period, and the result is interpreted as bacterial resistance against the tested concentration of an antimicrobial. In contrast, failed detection of bacterial biomass by MALDI-TOF MS defines inhibition of bacterial growth by an antibiotic in the tested concentration. The categorization of isolates as S, I or R was performed based on results for one or two breakpoint concentrations included for each antibiotic. The test was defined as valid, if the bacterial growth in control without an antibiotic was successfully detected by the MBT FAST prototype software.
The rates of categorical agreement (CA), very major error (VME, the number of false S results with DOT-MGA divided by the number of R results with standard method), major error (ME, the number of false R results with DOT-MGA divided by the number of S results with standard method) and minor error (mE, the number of false categorizations involving I category) were calculated for each antibiotic and each time point separately, as recommended by ISO [20] and FDA [21].
2.4. Broth Microdilution as Standard Method
After 6-µL samples were removed for DOT-MGA from the microtiter plates (MERLIN Diagnostika, Bornheim-Hersel, Germany) containing bacterial suspension and antibiotics, the incubation of these microtiter plates was performed for 18 ± 2 h at 35 ± 1 °C, as recommended by ISO [22] and CLSI [23] for broth microdilution method. The turbidity reading was performed visually, and the categorization results from the microtiter plate that was used for inoculation of the 18 h-DOT-MGA setup were used as reference for accuracy evaluation of the DOT-MGA.
Reference strains: E. coli ATCC 25922, E. coli ATCC 35218, K. pneumoniae ATCC 700603, K. pneumoniae ATCC BAA-1705 and Pseudomonas aeruginosa ATCC 27853 were used as quality control (QC).
3. Results
To evaluate the assay performance and compare it with the reference method, overall validity, categorical agreements (CA), very major errors (VME), major errors (ME) and minor errors (mE) were calculated for each antibiotic and each time point separately.
When the short incubation time of 6 h was implemented, the testing was valid (growth control detected) for all isolates and the overall CA was 92.0% (Table 1). With this incubation time, CA for individual antimicrobials varied between 70.8% (for cefepime) and 100% (for cefuroxime, cefotaxime, ceftazidime/avibactam, ertapenem, ciprofloxacin, levofloxacin, gentamicin, tobramycin and tigecycline). CA of ≥90% was achieved for 18 of 24 antimicrobials using this incubation duration.
After the 8-h incubation, all tests were valid, and the overall CA amounted to 92.7%. Applying this incubation duration, CA for individual antimicrobials ranged from 75.0% (for cefepime) to 100% (for cefuroxime, cefotaxime, ceftazidime/avibactam, ciprofloxacin, levofloxacin, moxifloxacin, gentamicin, tobramycin, trimethoprim/sulfamethoxazole and tigecycline). CA of ≥90% was achieved for 17 of 24 antimicrobials with the incubation for 8 h (Table 1).
Using the standard incubation time of 18 h, the test was valid (successful detection of growth control) in 75.2% of isolates and the CA calculated for valid tests was 90.5%. CA for individual antimicrobials varied between 77.8% (for ampicillin, cefepime and imipenem) and 100% (for cefotaxime, ceftazidime/avibactam, moxifloxacin, gentamicin, tobramycin and tigecycline). CA of ≥90% was achieved for 12 of 24 antimicrobials with the incubation for 18 h (Table 1).
4. Discussion
DOT-MGA is a novel MALDI-TOF MS-based AST method, which was reported to provide information on antimicrobial susceptibility or resistance in shorter time than the reference AST methods [11,12,13,14,16,17,18,19]. The assay principally relies on broth microdilution and is, therefore, a phenotypic growth-based method. Hence, potentially every antimicrobial can be tested with this universal method. Nevertheless, specific differences have been implemented in DOT-MGA, compared to the reference broth microdilution method. While the accuracy of the DOT-MGA has previously been demonstrated for several antimicrobials [12,16,17,18,19], the deviations from the standard method may hypothetically cause specific result deviation for particular antimicrobials not previously investigated, as it may be the case with every modified method. Such unknown effects may theoretically be caused, e.g., by the interference of antimicrobial substances with MALDI-TOF MS measurement, behavior of microorganisms in the presence of a specific antibiotic when incubated in microdroplets instead of 100-µL volume in a microtiter plate or microbial behavior on the surface of a MALDI target plate in the presence of a specific antibiotic. Therefore, this study aimed to test a wide range of antimicrobials from different classes in order to reveal any potential risk of inaccuracies with specific antimicrobials.
The short incubation time of 6 h resulted in an overall CA of 92.0% (Table 1), and the CA amounted to 92.7% after 8 h of incubation time. The overall CA after standard incubation time of 18 h was 90.5%. The individual performance of each antibiotic varied between 70.8% (for cefepime after 6 h incubation time) and 100% (for a large number of antibiotics after respective incubation time).
While the tested collection of consecutive clinical isolates reflected the occurrence in diagnostics routine, the number of resistant or susceptible strains was low for some antibiotics. Taking into account that the rates of VME and ME are calculated using the number of resistant or susceptible isolates, respectively, as denominator [20,21], the informative value of these parameters should be interpreted with caution (Table 1).
The best test performance was achieved when the short incubation times of six and eight hours were applied. The lower assay performance with the standard incubation time of 18 h is probably due to the fact that assay conditions and the evaluation algorithms were developed for a rapid test. With longer incubation, evaporation in microdroplets may become an issue that disturbs the microbial growth and the MALDI-TOF MS measurement. Indeed, a better control of humidity can be achieved with a dedicated incubation chamber than with the simple plastic box used in this study. Generally, this study was performed using “homebrew” utensils and a provisional software prototype.
Certainly, better control over the test conditions and, hence, better accuracy can be reached with standardized assay tools. The number of samples tested should also be increased and a broader range of resistance patterns tested to provide more reliable information on test performance. For the future, automated processing, standardized incubation tools and improved software analysis are needed that would enable comfortable workflow and increased test performance.
The MALDI-TOF MS-based DOT-MGA was shown in this proof-of-principle study to be a universal AST method suitable for rapid simultaneous testing of multiple antibiotics against clinical Enterobacterales isolates.
Supplementary Materials
The following is available online at https://www.mdpi.com/article/10.3390/diagnostics11101803/s1, Table S1: panel for simultaneous DOT-MGA testing of multiple antibiotics vs. Enterobacterales.
Author Contributions
E.A.I., I.D.N. and K.B. designed the experiments. I.D.N. and J.A.B. performed the experiments. E.A.I., I.D.N., K.S., O.D., M.K. and K.B. designed and analyzed specific MALDI-TOF MS instrument settings for experiments. I.D.N. performed initial data evaluation. E.A.I., I.D.N., K.S., O.D., M.K. and K.B. analyzed the data. E.A.I. wrote the manuscript with input from I.D.N., J.A.B., K.S., O.D., M.K. and K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by grants from the German Federal Ministry of Education and Research (BMBF) to E.A.I., K.B. (16GW0150) and M.K. (16GW0149K) and in part by the European Regional Development Fund (ERDF) to K.B. (grant number GHS-20-0010).
Data Availability Statement
Data are contained within the article or Supplementary Material.
Acknowledgments
We are grateful to Damayanti Kaiser, Michaela Schreiner, Annkatrin Bibo and Barbara Grünastel for their excellent technical assistance.
Conflicts of Interest
E.A.I. and K.B. are inventors of patent applications owned by the University of Münster and licensed to Bruker Daltonics GmbH & Co. KG, including a patent application on DOT-MGA. I.D.N., K.S., O.D. and M.K. are employees of Bruker Daltonics GmbH & Co. KG. J.A.B. declares no conflict of interest.
References
- Köck, R.; Wüllenweber, J.; Horn, D.; Lanckohr, C.; Becker, K.; Idelevich, E.A. Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob. Resist. Infect. Control. 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doern, G.V.; Vautour, R.; Gaudet, M.; Levy, B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 1994, 32, 1757–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zadka, H.; Raykhshtat, E.; Uralev, B.; Bishouty, N.; Weiss-Meilik, A.; Adler, A. The implementation of rapid microbial identification via MALDI-ToF reduces mortality in gram-negative but not gram-positive bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2053–2059. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Grunewald, C.M.; Wüllenweber, J.; Becker, K. Rapid identification and susceptibility testing of Candida spp. from positive blood cultures by combination of direct MALDI-TOF mass spectrometry and direct inoculation of Vitek 2. PLoS ONE 2014, 9, e114834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamy, B.; Sundqvist, M.; Idelevich, E.A. Bloodstream infections—Standard and progress in pathogen diagnostics. Clin. Microbiol. Infect. 2020, 26, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Becker, K. How to accelerate antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2019, 25, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Bachmann, T.T.; Lüdke, G.; Lisby, J.G.; Kahlmeter, G.; Mohess, A.; Becker, K.; Hays, J.P.; Woodford, N.; Mitsakakis, K.; et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2019, 17, 51–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burckhardt, I.; Zimmermann, S. Susceptibility Testing of Bacteria Using Maldi-Tof Mass Spectrometry. Front. Microbiol. 2018, 9, 1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oviaño, M.; Bou, G. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for the Rapid Detection of Antimicrobial Resistance Mechanisms and Beyond. Clin. Microbiol. Rev. 2018, 32, e00037-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bard, J.D.; Lee, F. Why Can’t We Just Use PCR? The Role of Genotypic versus Phenotypic Testing for Antimicrobial Resistance Testing. Clin. Microbiol. Newslett. 2018, 40, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Idelevich, E.A.; Becker, K. MALDI-TOF mass spectrometry for antimicrobial susceptibility testing. J. Clin. Microbiol. 2021, 01814–01819. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 2018, 24, 738–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neonakis, I.K.; Spandidos, D.A. MALDI-TOF mass spectrometry-based direct-on-target microdroplet growth assay: A novel assay for susceptibility testing and beyond. Future Microbiol. 2019, 14, 729–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neonakis, I.K.; Spandidos, D.A. MALDI-TOF MS-based direct-on-target microdroplet growth assay: Latest developments. Exp. Ther. Med. 2020, 20, 2555–2556. [Google Scholar] [CrossRef] [PubMed]
- EUCAST (2019) Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. Available online: http://www.eucast.org (accessed on 6 July 2021).
- Correa-Martínez, C.L.; Idelevich, E.A.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid Detection of Extended-Spectrum β-Lactamases (ESBL) and AmpC β-Lactamases in Enterobacterales: Development of a Screening Panel Using the MALDI-TOF MS-Based Direct-on-Target Microdroplet Growth Assay. Front. Microbiol. 2019, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Correa-Martínez, C.L.; Idelevich, E.A.; Sparbier, K.; Kuczius, T.; Kostrzewa, M.; Becker, K. Development of a MALDI-TOF MS-based screening panel for accelerated differential detection of carbapenemases in Enterobacterales using the direct-on-target microdroplet growth assay. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idelevich, E.A.; Storck, L.M.; Sparbier, K.; Drews, O.; Kostrzewa, M.; Becker, K. Rapid Direct Susceptibility Testing from Positive Blood Cultures by the Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry-Based Direct-on-Target Microdroplet Growth Assay. J. Clin. Microbiol. 2018, 56, e00913-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nix, I.D.; Idelevich, E.A.; Storck, L.M.; Sparbier, K.; Drews, O.; Kostrzewa, M.; Becker, K. Detection of Methicillin Resistance in Staphylococcus aureus From Agar Cultures and Directly From Positive Blood Cultures Using MALDI-TOF Mass Spectrometry-Based Direct-on-Target Microdroplet Growth Assay. Front. Microbiol. 2020, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- ISO (2007) 20776-2. Clinical Laboratory Testing and In Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 2: Evaluation of Performance of Antimicrobial Susceptibility Test Devices; International Organization for Standardization: Geneva, Switzerland, 2007. [Google Scholar]
- FDA. Guidance for Industry and FDA-Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems; US Food and Drug Administration: Silver Spring, MD, USA, 2009. [Google Scholar]
- ISO (2006) 20776-1. Clinical Laboratory Testing and In Vitro Diagnostic Test Systems-Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices-Part 1: Reference Method for Testing the in Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases; International Organization for Standardization: Geneva, Switzerland, 2006. [Google Scholar]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 11th ed. M07-A11; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).