Changes in Protein Structural Motifs upon Post-Translational Modification in Kidney Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Demography
2.2. Sample Preparation for MS Analysis
2.3. Mass Spectrometry Protein Registration
2.4. Protein Identification and Criteria Selection for PTMs
2.5. Molecular Dynamics Simulations
3. Results
3.1. Post-Translational Protein Modifications
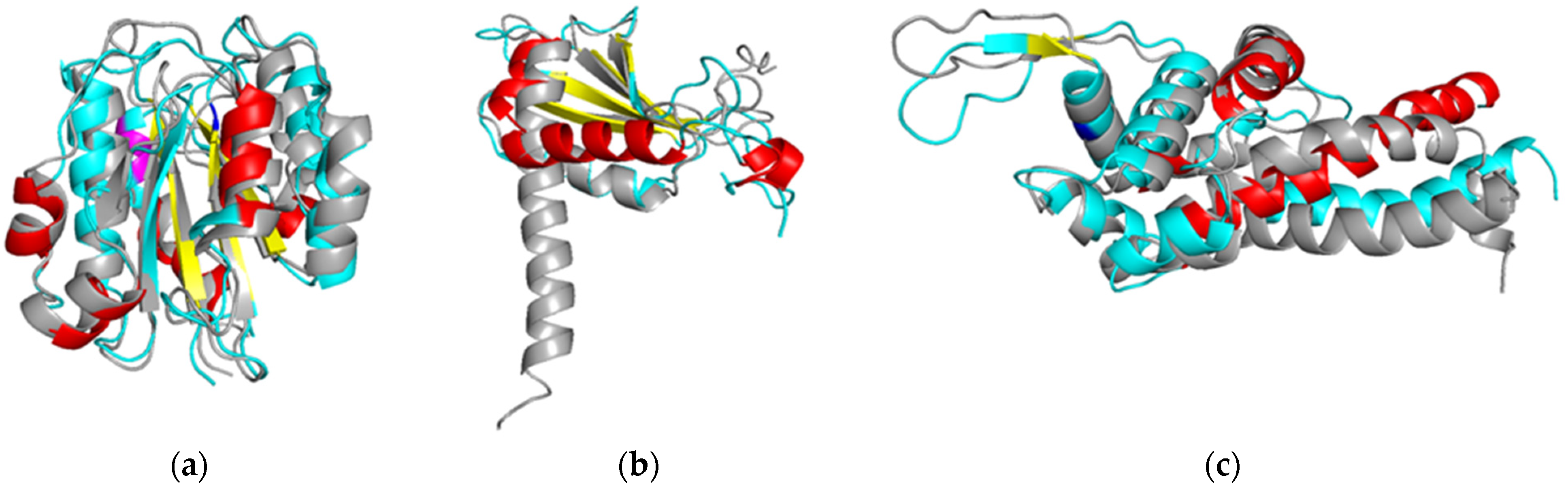
3.2. Structural Analysis of Proteins Carrying PTMs
3.3. Molecular Dynamics Simulation of Protein Molecules Containing PTMs Associated with Kidney Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-Omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [PubMed]
- Krug, K.; Mertins, P.; Zhang, B.; Hornbeck, P.; Raju, R.; Ahmad, R.; Szucs, M.; Mundt, F.; Forestier, D.; Jane-Valbuena, J.; et al. A Curated Resource for Phosphosite-Specific Signature Analysis. Mol. Cell. Proteom. 2019, 18, 576–593. [Google Scholar] [CrossRef]
- Mnatsakanyan, R.; Shema, G.; Basik, M.; Batist, G.; Borchers, C.H.; Sickmann, A.; Zahedi, R.P. Detecting Post-Translational Modification Signatures as Potential Biomarkers in Clinical Mass Spectrometry. Expert Rev. Proteom. 2018, 15, 515–535. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.S.; Prabhakaran, V.; Desai, A.P.; Bajpai, J.; Verma, R.J.; Swain, P.K. Post-Translational Modifications (PTMs), from a Cancer Perspective: An Overview. Oncogen 2019, 2, 12. [Google Scholar] [CrossRef]
- Díaz-Fernández, A.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Post-Translational Modifications in Tumor Biomarkers: The next Challenge for Aptamers? Anal. Bioanal. Chem. 2018, 410, 2059–2065. [Google Scholar] [CrossRef]
- Heo, K.-S. Regulation of Post-Translational Modification in Breast Cancer Treatment. BMB Rep. 2019, 52, 113–118. [Google Scholar] [CrossRef]
- Zou, X.; Blank, M. Targeting P38 MAP Kinase Signaling in Cancer through Post-Translational Modifications. Cancer Lett. 2017, 384, 19–26. [Google Scholar] [CrossRef]
- Celano, M.; Mio, C.; Sponziello, M.; Verrienti, A.; Bulotta, S.; Durante, C.; Damante, G.; Russo, D. Targeting Post-Translational Histone Modifications for the Treatment of Non-Medullary Thyroid Cancer. Mol. Cell. Endocrinol. 2018, 469, 38–47. [Google Scholar] [CrossRef]
- Shi, A.-M.; Tao, Z.-Q.; Li, R.; Wang, Y.-Q.; Wang, X.; Zhao, J. Vimentin and Post-Translational Modifications in Cell Motility during Cancer—A Review. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2603–2606. [Google Scholar]
- Perri, A.M.; Agosti, V.; Olivo, E.; Concolino, A.; Angelis, M.D.; Tammè, L.; Fiumara, C.V.; Cuda, G.; Scumaci, D. Histone Proteomics Reveals Novel Post-Translational Modifications in Breast Cancer. Aging 2019, 11, 11722–11755. [Google Scholar] [CrossRef]
- Cooper, A.; Woulfe, D.; Kilic, F. Post-Translational Modifications of Serotonin Transporter. Pharmacol. Res. 2019, 140, 7–13. [Google Scholar] [CrossRef]
- Wende, A.R. Post-translational Modifications of the Cardiac Proteome in Diabetes and Heart Failure. Proteom.—Clin. Appl. 2016, 10, 25–38. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today/home (accessed on 8 September 2021).
- Kopylov, A.T.; Petrovsky, D.V.; Stepanov, A.A.; Rudnev, V.R.; Malsagova, K.A.; Butkova, T.V.; Zakharova, N.V.; Kostyuk, G.P.; Kulikova, L.I.; Enikeev, D.V.; et al. Convolutional Neural Network in Proteomics and Metabolomics for Determination of Comorbidity between Cancer and Schizophrenia. J. Biomed. Inform. 2021, 122, 103890. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Tikhonov, D.A.; Kulikova, L.I.; Efimov, A.V. The Study of Interhelical Angles in the Structural Motifs Formed by Two Helices. Math. Biol. Bioinform. 2019, 14, t1–t17. [Google Scholar] [CrossRef]
- Tikhonov, D.A.; Kulikova, L.I.; Efimov, A.V. Analysis of the Torsion Angles between Helical Axes in Pairs of Helices in Protein Molecules. Math. Biol. Bioinform. 2018, 13, t17–t28. [Google Scholar] [CrossRef]
- Tikhonov, D.A.; Kulikova, L.I.; Efimov, A.V. Statistical Analysis of the Internal Distances of Helical Pairs in Protein Molecules. Math. Biol. Bioinform. 2019, 14, t18–t36. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Petrov, D.; Margreitter, C.; Grandits, M.; Oostenbrink, C.; Zagrovic, B. A Systematic Framework for Molecular Dynamics Simulations of Protein Post-Translational Modifications. PLoS Comput. Biol. 2013, 9, e1003154. [Google Scholar] [CrossRef] [PubMed]
- Margreitter, C.; Petrov, D.; Zagrovic, B. Vienna-PTM Web Server: A Toolkit for MD Simulations of Protein Post-Translational Modifications. Nucleic Acids Res. 2013, 41, W422–W426. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.; Mark, A.E.; Peggion, E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 27–28. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Modification of the Generalized Born Model Suitable for Macromolecules. J. Phys. Chem. B 2000, 104, 3712–3720. [Google Scholar] [CrossRef]
- Rudnev, V.R.; Kulikova, L.I.; Kaysheva, A.L.; Efimov, A.V.; Tikhonov, D.A. Use of the Molecular Dynamics Method to Investigate the Stability of α-α-Corner Structural Motifs in Proteins. Symmetry 2021, 13, 1193. [Google Scholar] [CrossRef]
- Tikhonov, D.; Kulikova, L.; Kopylov, A.; Malsagova, K.; Stepanov, A.; Rudnev, V.; Kaysheva, A. Super Secondary Structures of Proteins with Post-Translational Modifications in Colon Cancer. Molecules 2020, 25, 3144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, W. Protein Post-Translational Modifications in Head and Neck Cancer. Front. Oncol. 2020, 10, 571944. [Google Scholar] [CrossRef] [PubMed]
- Doyle, H.A.; Mamula, M.J. Post-Translational Protein Modifications in Antigen Recognition and Autoimmunity. Trends Immunol. 2001, 22, 443–449. [Google Scholar] [CrossRef]
- Wang, J.; Yang, L.; Liang, F.; Chen, Y.; Yang, G. Integrin Alpha x Stimulates Cancer Angiogenesis through PI3K/Akt Signaling–Mediated VEGFR2/VEGF-A Overexpression in Blood Vessel Endothelial Cells. J. Cell. Biochem. 2019, 120, 1807–1818. [Google Scholar] [CrossRef]
- Sui, Y.; Lu, K.; Fu, L. Prediction and Analysis of Novel Key Genes ITGAX, LAPTM5, SERPINE1 in Clear Cell Renal Cell Carcinoma through Bioinformatics Analysis. PeerJ 2021, 9, e11272. [Google Scholar] [CrossRef]
- Shi, D.; Zhong, Z.; Xu, R.; Li, B.; Li, J.; Habib, U.; Peng, Y.; Mao, H.; Li, Z.; Huang, F.; et al. Association of ITGAX and ITGAM Gene Polymorphisms with Susceptibility to IgA Nephropathy. J. Hum. Genet. 2019, 64, 927–935. [Google Scholar] [CrossRef]
- Mimura, I.; Tojo, A.; Kinugasa, S.; Uozaki, H.; Fujita, T. Renal Cell Carcinoma in Association with IgA Nephropathy in the Elderly. Am. J. Med. Sci. 2009, 338, 431–432. [Google Scholar] [CrossRef]
- Willmann, K.L.; Milosevic, S.; Pauklin, S.; Schmitz, K.-M.; Rangam, G.; Simon, M.T.; Maslen, S.; Skehel, M.; Robert, I.; Heyer, V.; et al. A Role for the RNA Pol II-Associated PAF Complex in AID-Induced Immune Diversification. J. Exp. Med. 2012, 209, 2099–2111. [Google Scholar] [CrossRef]
- Fang, Z.-P.; Jiang, B.-G.; Zhang, F.-B.; Wang, A.-D.; Ji, Y.-M.; Xu, Y.-F.; Li, J.-C.; Zhou, W.-P.; Zhou, W.-J.; Han, H.-X. Rpb3 Promotes Hepatocellular Carcinoma through Its N-Terminus. Oncotarget 2014, 5, 9256–9268. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, Q.; Xu, J.; Zhao, W.; Zhang, Z.; Xu, D.; Zhang, Y.; Zhao, E.; Zhao, G. Somatic Mutation of DNAH Genes Implicated Higher Chemotherapy Response Rate in Gastric Adenocarcinoma Patients. J. Transl. Med. 2019, 17, 109. [Google Scholar] [CrossRef]
- García-Mata, R.; Bebök, Z.; Sorscher, E.J.; Sztul, E.S. Characterization and Dynamics of Aggresome Formation by a Cytosolic GFP-Chimera. J. Cell Biol. 1999, 146, 1239–1254. [Google Scholar] [CrossRef]
- Ambudkar, S.V. Drug-Stimulatable ATPase Activity in Crude Membranes of Human MDR1-Transfected Mammalian Cells. Methods Enzymol. 1998, 292, 504–514. [Google Scholar] [CrossRef]
- Domenichini, A.; Adamska, A.; Falasca, M. ABC Transporters as Cancer Drivers: Potential Functions in Cancer Development. Biochim. Biophys. Acta BBA—Gen. Subj. 2019, 1863, 52–60. [Google Scholar] [CrossRef]
- Begicevic, R.-R.; Falasca, M. ABC Transporters in Cancer Stem Cells: Beyond Chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef]
- Aye, I.L.M.H.; Singh, A.T.; Keelan, J.A. Transport of Lipids by ABC Proteins: Interactions and Implications for Cellular Toxicity, Viability and Function. Chem. Biol. Interact. 2009, 180, 327–339. [Google Scholar] [CrossRef]
- Pasello, M.; Giudice, A.M.; Scotlandi, K. The ABC Subfamily A Transporters: Multifaceted Players with Incipient Potentialities in Cancer. Semin. Cancer Biol. 2020, 60, 57–71. [Google Scholar] [CrossRef]
- Xia, C.; Ma, W.; Stafford, L.J.; Liu, C.; Gong, L.; Martin, J.F.; Liu, M. GGAPs, a New Family of Bifunctional GTP-Binding and GTPase-Activating Proteins. Mol. Cell. Biol. 2003, 23, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-Y.; Ye, K. PIKE GTPase Signaling and Function. Int. J. Biol. Sci. 2005, 1, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Snyder, S.H. PIKE GTPase: A Novel Mediator of Phosphoinositide Signaling. J. Cell Sci. 2004, 117, 155–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ha, V.L.; Luo, R.; Nie, Z.; Randazzo, P.A. Contribution of AZAP-Type Arf GAPs to Cancer Cell Migration and Invasion. Adv. Cancer Res. 2008, 101, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Hurt, K.J.; Wu, F.Y.; Fang, M.; Luo, H.R.; Hong, J.J.; Blackshaw, S.; Ferris, C.D.; Snyder, S.H. PIKE: A Nuclear Gtpase That Enhances PI3kinase Activity and Is Regulated by Protein 4.1N. Cell 2000, 103, 919–930. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, Q.; Luh, F.; Jin, B.; Liu, X. Overexpression of the ASPM Gene Is Associated with Aggressiveness and Poor Outcome in Bladder Cancer. Oncol. Lett. 2019, 17, 1865–1876. [Google Scholar] [CrossRef]
- Levine, A.J.; Puzio-Kuter, A.M. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef]
- Georgila, K.; Vyrla, D. Apolipoprotein A-I (ApoA-I), Immunity, Inflammation and Cancer. Cancers 2019, 11, 1097. [Google Scholar] [CrossRef]
- Chinese Human Proteome Project (CNHPP) Consortium; Jiang, Y.; Sun, A.; Zhao, Y.; Ying, W.; Sun, H.; Yang, X.; Xing, B.; Sun, W.; Ren, L.; et al. Proteomics Identifies New Therapeutic Targets of Early-Stage Hepatocellular Carcinoma. Nature 2019, 567, 257–261. [Google Scholar] [CrossRef]
- Guo, S.; He, X.; Chen, Q.; Yang, G.; Yao, K.; Dong, P.; Ye, Y.; Chen, D.; Zhang, Z.; Qin, Z.; et al. The Effect of Preoperative Apolipoprotein A-I on the Prognosis of Surgical Renal Cell Carcinoma: A Retrospective Large Sample Study. Medicine 2016, 95, e3147. [Google Scholar] [CrossRef]
- Liu, Z.; Xiao, Y.; Tang, L.; Jiang, L.; Wang, Y.; Zhang, R.; Wei, Q.; Lu, Y. Apolipoprotein A1 −75 G/A and +83 C/T Polymorphisms and Renal Cancer Risk. Lipids Health Dis. 2015, 14, 143. [Google Scholar] [CrossRef]
- Luza, S.C.; Speisky, H.C. Liver Copper Storage and Transport during Development: Implications for Cytotoxicity. Am. J. Clin. Nutr. 1996, 63, 812S–820S. [Google Scholar] [CrossRef]
- Kondi-Pafiti, A.; Smyrniotis, V.; Frangou, M.; Papayanopoulou, A.; Englezou, M.; Deligeorgi, H. Immunohistochemical Study of Ceruloplasmin, Lactoferrin and Secretory Component Expression in Neoplastic and Non-Neoplastic Thyroid Gland Diseases. Acta Oncol. 2000, 39, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.; Teknos, T.N.; Barrios, M.; Brewer, G.J.; Dick, R.D.; Merajver, S.D. The Role of Copper Suppression as an Antiangiogenic Strategy in Head and Neck Squamous Cell Carcinoma. Laryngoscope 2001, 111, 696–701. [Google Scholar] [CrossRef]
- Allison, S.J. PAX8: A Candidate Oncogene in RCC. Nat. Rev. Nephrol. 2019, 15, 662. [Google Scholar] [CrossRef] [PubMed]
- Bleu, M.; Gaulis, S.; Lopes, R.; Sprouffske, K.; Apfel, V.; Holwerda, S.; Pregnolato, M.; Yildiz, U.; Cordo’, V.; Dost, A.F.M.; et al. PAX8 Activates Metabolic Genes via Enhancer Elements in Renal Cell Carcinoma. Nat. Commun. 2019, 10, 3739. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, G.; Taskén, K. Specificity and Spatial Dynamics of Protein Kinase A Signaling Organized by A-Kinase-Anchoring Proteins. J. Mol. Endocrinol. 2010, 44, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Dodge-Kafka, K.L.; Bauman, A.; Kapiloff, M.S. A-Kinase Anchoring Proteins as the Basis for CAMP Signaling. Handb. Exp. Pharmacol. 2008, 186, 3–14. [Google Scholar] [CrossRef]
- Chen, L.; Marquardt, M.L.; Tester, D.J.; Sampson, K.J.; Ackerman, M.J.; Kass, R.S. Mutation of an A-Kinase-Anchoring Protein Causes Long-QT Syndrome. Proc. Natl. Acad. Sci. USA 2007, 104, 20990–20995. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-Y.; Liu, Y.-P.; Xie, L.-Y.; Wang, X.-Y.; Yang, F.; Chen, S.-Y.; Li, Z.-G. AKAP-9 Promotes Colorectal Cancer Development by Regulating Cdc42 Interacting Protein 4. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2016, 1862, 1172–1181. [Google Scholar] [CrossRef]
- Frank, B.; Wiestler, M.; Kropp, S.; Hemminki, K.; Spurdle, A.B.; Sutter, C.; Wappenschmidt, B.; Chen, X.; Beesley, J.; Hopper, J.L.; et al. Association of a Common AKAP9 Variant with Breast Cancer Risk: A Collaborative Analysis. JNCI J. Natl. Cancer Inst. 2008, 100, 437–442. [Google Scholar] [CrossRef]
- Ciampi, R.; Knauf, J.A.; Kerler, R.; Gandhi, M.; Zhu, Z.; Nikiforova, M.N.; Rabes, H.M.; Fagin, J.A.; Nikiforov, Y.E. Oncogenic AKAP9-BRAF Fusion Is a Novel Mechanism of MAPK Pathway Activation in Thyroid Cancer. J. Clin. Investig. 2005, 115, 94–101. [Google Scholar] [CrossRef]
- Herter, J.M.; Grabie, N.; Cullere, X.; Azcutia, V.; Rosetti, F.; Bennett, P.; Herter-Sprie, G.S.; Elyaman, W.; Luscinskas, F.W.; Lichtman, A.H.; et al. AKAP9 Regulates Activation-Induced Retention of T Lymphocytes at Sites of Inflammation. Nat. Commun. 2015, 6, 10182. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Powers, A.D.; Palecek, S.P. Protein Analytical Assays for Diagnosing, Monitoring, and Choosing Treatment for Cancer Patients. J. Healthc. Eng. 2012, 3, 503–534. [Google Scholar] [CrossRef]
- Chandler, K.; Goldman, R. Glycoprotein Disease Markers and Single Protein-Omics. Mol. Cell. Proteom. 2013, 12, 836–845. [Google Scholar] [CrossRef] [PubMed]

| Sequence with PTM | Gene Name | Protein Name | Frequency, % |
|---|---|---|---|
| SPAGPAATPAQAQAAS-P(T)-PRK | TCOF | Treacle protein | 56 |
| QE-AC(K)-EQVSLR | AKAP9 | A-kinase anchor protein 9 | 49 |
| ILIVI-P(T)-DGK | ITAX | Integrin alpha-X | 37 |
| LEPIA-P(T)-EVWLINK | ABCA1 | ATP-binding cassette sub-Family A, Member 1 | 35 |
| LSAQA-P(S)-LKR | JKIP1 | Janus kinase and microtubule-interacting protein 1 | 35 |
| Y-AC(K)-DPVTVVVDDLR | ACAP1 | Arf-GAP with coiled-coil | 33 |
| LYLAVKNAN-AC(K) | ASPM | Abnormal spindle-like microcephaly-associated protein | 30 |
| IEDFWGPA-AC(K)-R | DYH7 | Dynein heavy chain 7 | 23 |
| LILEHQE-AC(K) | SCLT1 | Sodium channel and clathrin linker 1 | 12 |
| IK-P(Y)-APISGGDHAEVDVPK | TENA | Tenascin | 7 |
| P(Y)-HWEHTGLTLR | APOB | Apolipoprotein B-100 | 5 |
| VPHPLEH-AC(K)-IIIR | RPB1B | DNA-directed RNA polymerase II, Subunit RPB11-a | 5 |
| AC(K)-WQEEMELYR | APOA1 | Apolipoprotein A-I | 2 |
| VTFHN-AC(K)-GAYPLSIEPIGVR | CERU | Ceruloplasmin | 2 |
| LLLT-P(T)-QFPFKINEK | ORC3 | Origin recognition complex Subunit 3 | 2 |
| Gene Name | Nprot | VN | VM | UN | UM | LOC | PTM SEQQ | PROTEIN |
|---|---|---|---|---|---|---|---|---|
| ITAX | 5 | 3.92 | 4.12 | 49.82 | 48.54 | T6 | ILIVITDGK | Integrin alpha-X |
| ABCA1 | 1 | 47.5 | 49.5 | 409.7 | 408.6 | T6 | LEPIATEVWLINK | ATP-binding cassette Sub-Family A, Member 1 |
| ACAP1 | 10 | 170.09 | 222.4 | 352.68 | 321.39 | K2 | YKDPVTVVVDDLR | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 |
| DYH7 | 1 | 88.7 | 123.4 | 304.8 | 321.6 | K9 | IEDFWGPAKR | Dynein heavy chain 7, axonemal |
| RPB1B | 13 | 76.215 | 121.877 | 269.138 | 297.762 | K8 | VPHPLEHKIIIR | DNA-directed RNA polymerase II subunit RPB11-a |
| APOA1 | 24 | 87.683 | 131.825 | 299.1625 | 326.867 | K1 | KWQEEMELYR | Apolipoprotein A-I |
| CERU | 4 | 91.95 | 110.725 | 246.475 | 247.825 | K6 | VTFHNKGAYPLSIEPIGVR | Ceruloplasmin |
| ORC3 | 5 | 41.34 | 88.46 | 390.64 | 396,9 | T5 | LLLTTQFPFKINEK | Origin recognition complex, Subunit 3 |
| Gene Name | PTM SEQQ | Motif 1 | Motif 2 | Motif 3 | Motif 4 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Locus | Nhelix | Locus | Nhelix | Locus | Nhelix | Locus | Nhelix | ||
| DYH7 | IEDFWGPA-AC(K)-R | 44–110 HA (44–57) HB (104–110) | 4 | 44–128 HA (44–57) HB (122–128) | 5 | 44–86 HA (44–57) HB (77–86) | 2 | 77–96 HA (44–57) HB (89–96) | 2 |
| ITAX | ILIVI-P(T)-DGK | 81–132 HA (305–316) HB (452–465) | 2 | 81–157 HA (305–316) HB (523–527) | 3 | 12–188 HA (81–90) HB (179–188) | 6 | – | – |
| ABCA1 | LEPIA-P(T)-EVWLINK | 305–465 HA (81–90) HB (125–132) | 8 | 305–527 HA (81–90) HB (125–132) | 9 | 326–545 HA (326–341) HB (529–545) | 9 | – | – |
| ORC3 | LLLT-P(T)-QFPFKINEK | 205–241 HA (205–216) HB (228–241) | 2 | 205–261 HA (205–216) HB (245–261) | 3 | 205–368 HA (205–216) HB (359–368) | 9 | – | – |
| RPB1B | VPHPLEH-AC(K)-IIIR | 40–112 HA (40–57) HB (83–112) | 2 | – | – | – | – | – | – |
| CERU | VTFHN-AC(K)-GAYPLSIEPIGVR | 322–516 HA (322–327) HB (510–516) | 3 | 377–565 HA (377–382) HB (557–565) | 3 | 377–672 HA (377–382) HB (667–672) | 5 | 377–856 HA (377–382) HB (851–856) | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tikhonov, D.; Kulikova, L.; Rudnev, V.; Kopylov, A.T.; Taldaev, A.; Stepanov, A.; Malsagova, K.; Izotov, A.; Enikeev, D.; Potoldykova, N.; et al. Changes in Protein Structural Motifs upon Post-Translational Modification in Kidney Cancer. Diagnostics 2021, 11, 1836. https://doi.org/10.3390/diagnostics11101836
Tikhonov D, Kulikova L, Rudnev V, Kopylov AT, Taldaev A, Stepanov A, Malsagova K, Izotov A, Enikeev D, Potoldykova N, et al. Changes in Protein Structural Motifs upon Post-Translational Modification in Kidney Cancer. Diagnostics. 2021; 11(10):1836. https://doi.org/10.3390/diagnostics11101836
Chicago/Turabian StyleTikhonov, Dmitry, Liudmila Kulikova, Vladimir Rudnev, Arthur T. Kopylov, Amir Taldaev, Alexander Stepanov, Kristina Malsagova, Alexander Izotov, Dmitry Enikeev, Natalia Potoldykova, and et al. 2021. "Changes in Protein Structural Motifs upon Post-Translational Modification in Kidney Cancer" Diagnostics 11, no. 10: 1836. https://doi.org/10.3390/diagnostics11101836
APA StyleTikhonov, D., Kulikova, L., Rudnev, V., Kopylov, A. T., Taldaev, A., Stepanov, A., Malsagova, K., Izotov, A., Enikeev, D., Potoldykova, N., & Kaysheva, A. (2021). Changes in Protein Structural Motifs upon Post-Translational Modification in Kidney Cancer. Diagnostics, 11(10), 1836. https://doi.org/10.3390/diagnostics11101836







