Deep Semi-Supervised Algorithm for Learning Cluster-Oriented Representations of Medical Images Using Partially Observable DICOM Tags and Images
Abstract
:1. Introduction
- We propose a method for exploiting DICOM tag information to construct pairwise relations using the Gower distance. After the Gower distance is calculated, thresholding is applied to create must-link and cannot-link pairwise constraints. By using this distance, we address the issue of missing data as well as the heterogeneity of data types across features.
- Our method is not limited to data having a single target value. Instead, it can be used on data where each image can be described using multiple target variables, i.e., DICOM tags.
- To introduce pairwise information during training, we propose a cost function where, along with the classical deep embedded clustering (DEC) loss and the reconstruction loss, we minimise the Kullback–Leibler (KL) divergence between the distributions of instances belonging to the same cluster, while also maximising the KL divergence for the pairs not belonging to the same cluster.
- We compare our model against the unsupervised convolutional improved deep embedded clustering (IDEC) model and with the semi-supervised algorithms combined with the popular feature descriptors. Results show that using additional DICOM tags can improve the clustering performance.
- We show that the model generalises well by observing the two-dimensional t-SNE of the feature embedding space, calculated over a disjoint test set.
2. Related Work
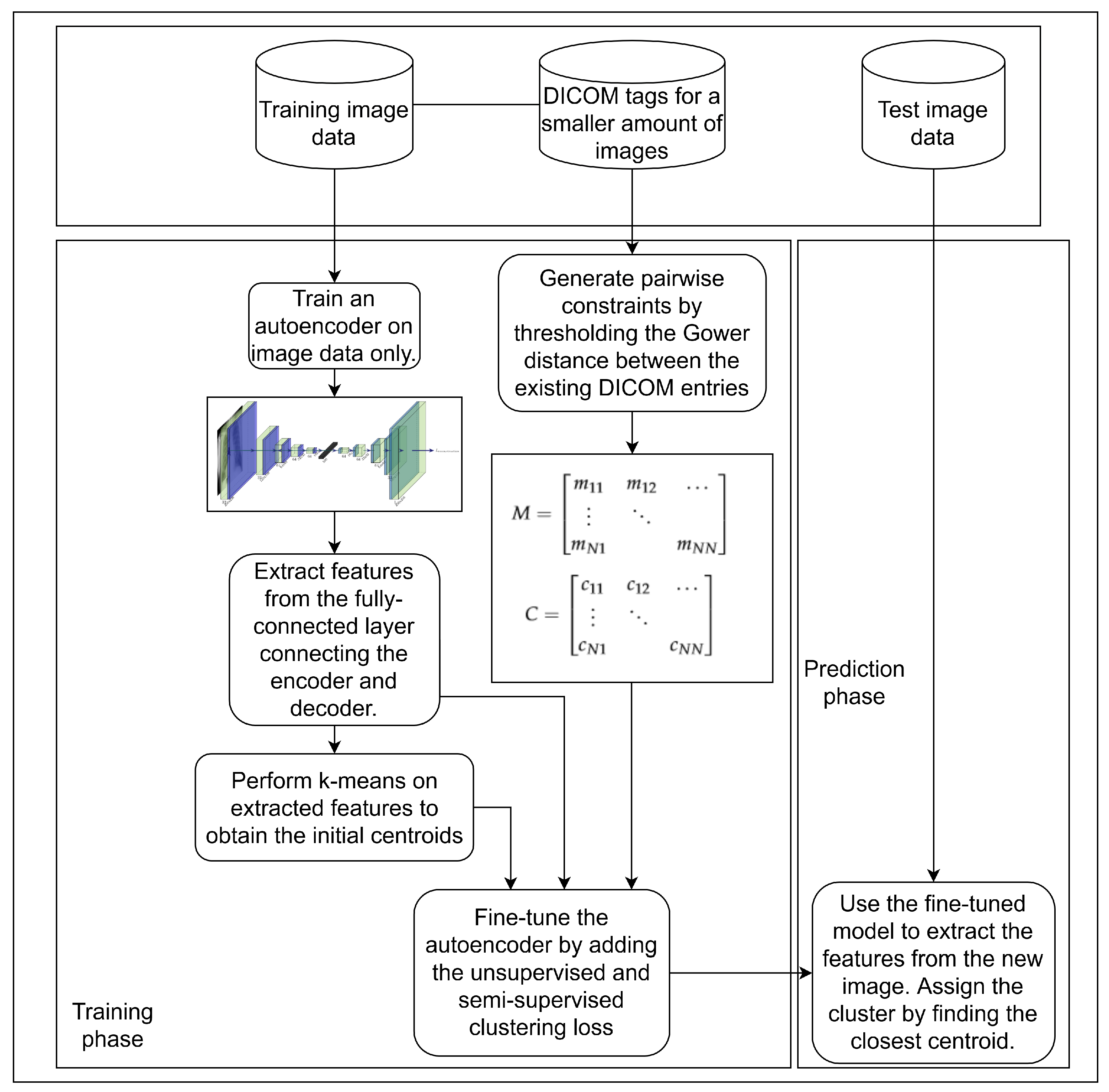
3. Materials and Methods
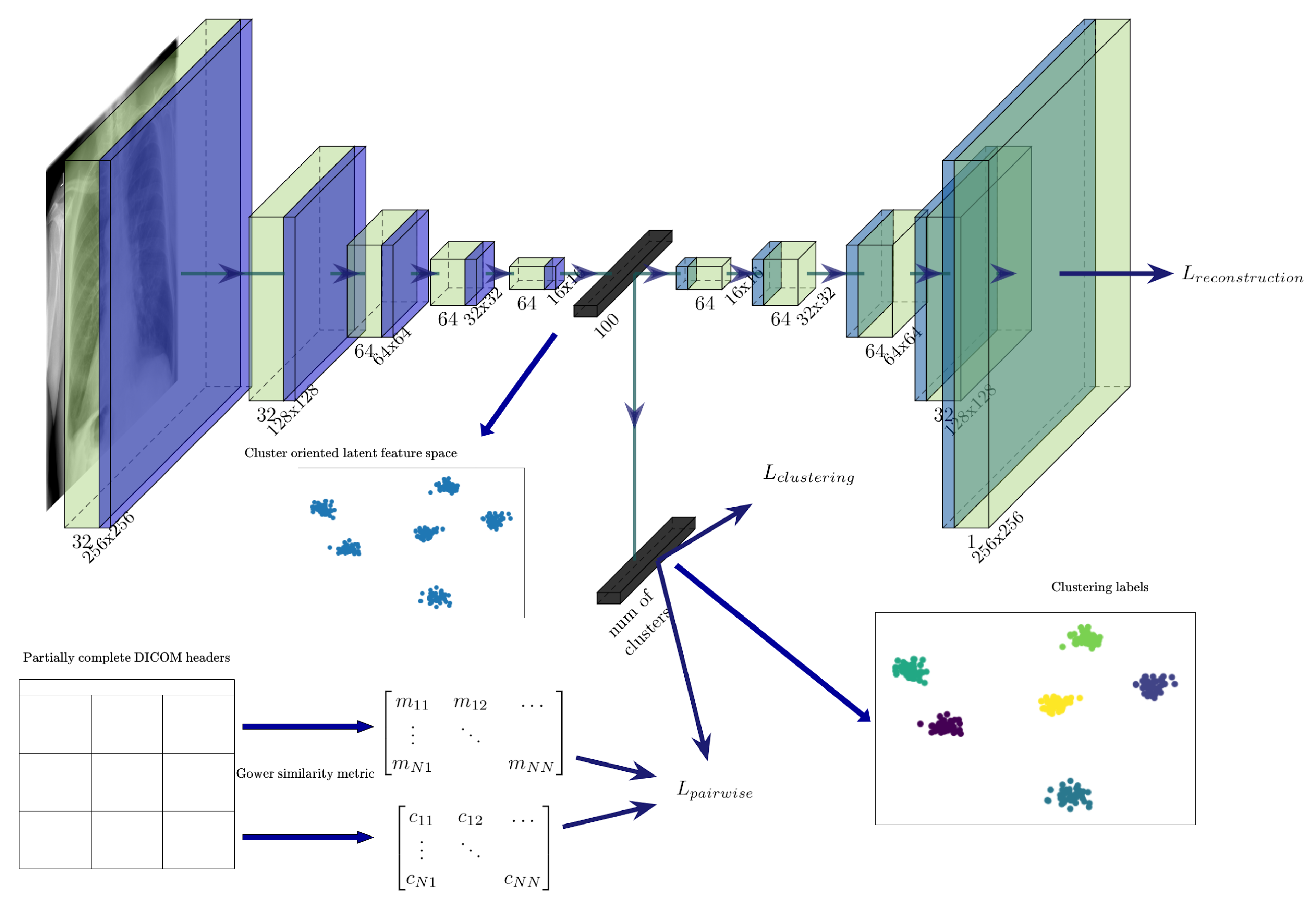
3.1. Unsupervised Pretraining of a Feature Extractor on Images
3.2. Using Gower Distance to Define Pairwise Constraints
3.3. Semi-Supervised Clustering with Pairwise Constraints
| Algorithm 1 Semi-supervised model-training algorithm utilising DICOM tags and images |
Require: Dataset (images coupled with DICOM tags, where available), number of clusters K, weights for the loss function (, , ), and for Gower distance used to calculate must-link and cannot-link pairwise relations, threshold for stopping the training, , .
|
3.4. Dataset
3.5. Model Evaluation and Experimental Setup
4. Results
5. Discussion
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PACS | Picture Archiving and Communication System |
| DICOM | Digital Imaging and Communications in Medicine |
| CAE | Convolutional autoencoder |
| MSE | Mean squared error |
| DEC | Deep embedded clustering |
| IDEC | Improved deep embedded clustering |
| PC k-means | Pairwise constrained k-means |
| KL | Kullback–Leibler |
| NMI | Normalised mutual information |
| HS | Homogeneity score |
| PCA | Principal component analysis |
| HOG | Histogram of oriented gradients |
| LBP | Local binary pattern |
| CHC | Clinical Hospital Centre |
| Mod | Modality |
| BPE | Body Part Examined |
| AR | Anatomic region |
| CT | Computed tomography |
| XA | X-ray angiography |
| NM | Nuclear medicine |
| RF | Radio fluoroscopy |
| MR | Magnetic resonance |
| CR | Computed radiography |
References
- Bidgood, W.D.; Horii, S.C.; Prior, F.W.; Van Syckle, D.E. Understanding and Using DICOM, the Data Interchange Standard for Biomedical Imaging. J. Am. Med. Inform. Assoc. 1997, 4, 199–212. [Google Scholar] [CrossRef] [Green Version]
- Dimitrovski, I.; Kocev, D.; Loskovska, S.; Džeroski, S. Hierarchical annotation of medical images. Pattern Recognit. 2011, 44, 2436–2449. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Källman, H.E.; Halsius, E.; Olsson, M.; Stenström, M. DICOM metadata repository for technical information in digital medical images. Acta Oncol. 2009, 48, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Bhattacharya, P.; Desai, B.C. A Framework for Medical Image Retrieval Using Machine Learning and Statistical Similarity Matching Techniques with Relevance Feedback. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Gueld, M.O.; Kohnen, M.; Keysers, D.; Schubert, H.; Wein, B.B.; Bredno, J.; Lehmann, T.M. Quality of DICOM header information for image categorization. In Medical Imaging 2002: PACS and Integrated Medical Information Systems: Design and Evaluation; Siegel, E.L., Huang, H.K., Eds.; International Society for Optics and Photonics, SPIE: San Diego, CA, USA, 2002; Volume 4685, pp. 280–287. [Google Scholar] [CrossRef]
- Manojlović, T.; Ilić, D.; Miletić, D.; Štajduhar, I. Using DICOM Tags for Clustering Medical Radiology Images into Visually Similar Groups. In Proceedings of the 9th International Conference on Pattern Recognition Applications and Methods, ICPRAM 2020, Valletta, Malta, 22–24 February 2020; Marsico, M.D., di Baja, G.S., Fred, A.L.N., Eds.; SCITEPRESS: Setúbal, Portugal, 2020; pp. 510–517. [Google Scholar] [CrossRef]
- Gauriau, R.; Bridge, C.; Chen, L.; Kitamura, F.; Tenenholtz, N.; Kirsch, J.; Andriole, K.; Michalski, M.; Bizzo, B. Using DICOM Metadata for Radiological Image Series Categorization: A Feasibility Study on Large Clinical Brain MRI Datasets. J. Digit. Imaging 2020, 33, 747–762. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.; Rudrapatna, M.; Sowmya, A. Automatic Lung Segmentation: A Comparison of Anatomical and Machine Learning Approaches. In Proceedings of the 2004 Intelligent Sensors, Sensor Networks and Information Processing Conference, Melbourne, Australia, 14–17 December 2004; pp. 451–456. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, T.M.; Schubert, H.; Keysers, D.; Kohnen, M.; Wein, B.B. The IRMA code for unique classification of medical images. In Proceedings of the Medical Imaging 2003: PACS and Integrated Medical Information Systems: Design and Evaluation, San Diego, CA, USA, 15–20 February 2003; Volume 5033, p. 440. [Google Scholar] [CrossRef] [Green Version]
- Wagstaff, K.L.; Cardie, C. Clustering with Instance-level Constraints. In Proceedings of the 17th International Conference on Machine Learning, Standord, CA, USA, 29 June–2 July 2000; pp. 1103–1110. [Google Scholar]
- Wagstaff, K.; Cardie, C.; Rogers, S.; Schrödl, S. Constrained K-means Clustering with Background Knowledge. In Proceedings of the International Conference on Machine Learning ICML, Williamstown, MA, USA, 28 June–1 July 2001; pp. 577–584. [Google Scholar]
- Basu, S.; Banerjee, A.; Mooney, R.J. Active semi-supervision for pairwise constrained clustering. In Proceedings of the 2004 SIAM International Conference on Data Mining (SDM), Lake Buena Vista, FL, USA, 22–24 April 2004. [Google Scholar] [CrossRef] [Green Version]
- Kohonen, T. The self-organizing map. Proc. IEEE 1990, 78, 1464–1480. [Google Scholar] [CrossRef]
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A Survey of Clustering With Deep Learning: From the Perspective of Network Architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Ng, A.Y.; Jordan, M.I.; Weiss, Y. On Spectral Clustering: Analysis and an algorithm. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2001; pp. 849–856. [Google Scholar]
- Xie, J.; Girshick, R.; Farhadi, A. Unsupervised Deep Embedding for Clustering Analysis. In Proceedings of the 33rd International Conference on Machine Learning; Balcan, M.F., Weinberger, K.Q., Eds.; PMLR: New York, NY, USA, 2016; Volume 48, pp. 478–487. [Google Scholar]
- Manojlovic, T.; Milanic, M.; Stajduhar, I. Deep embedded clustering algorithm for clustering PACS repositories. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Kira, Z. Neural network-based clustering using pairwise constraints. arXiv 2015, arXiv:1511.06321. [Google Scholar]
- Ren, Y.; Hu, K.; Dai, X.; Pan, L.; Hoi, S.C.; Xu, Z. Semi-supervised deep embedded clustering. Neurocomputing 2019, 325, 121–130. [Google Scholar] [CrossRef]
- Tian, T.; Zhang, J.; Lin, X.; Wei, Z.; Hakonarson, H. Model-based deep embedding for constrained clustering analysis of single cell RNA-seq data. Nat. Commun. 2021, 12, 1873. [Google Scholar] [CrossRef] [PubMed]
- Enguehard, J.; O’Halloran, P.; Gholipour, A. Semi-Supervised Learning with Deep Embedded Clustering for Image Classification and Segmentation. IEEE Access 2019, 7, 11093–11104. [Google Scholar] [CrossRef]
- Śmieja, M.; Struski, Ł.; Figueiredo, M.A. A classification-based approach to semi-supervised clustering with pairwise constraints. Neural Netw. 2020, 127, 193–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Basu, S.; Davidson, I. A Framework for Deep Constrained Clustering—Algorithms and Advances. In Machine Learning and Knowledge Discovery in Databases; Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2020; pp. 57–72. [Google Scholar] [CrossRef]
- Masci, J.; Meier, U.; Cireşan, D.; Schmidhuber, J. Stacked convolutional auto-encoders for hierarchical feature extraction. In Artificial Neural Networks and Machine Learning—ICANN 2011; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2011; pp. 52–59. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Shi, X.; Zhang, Y.; Wu, D.; Guizani, M. Deep Features Learning for Medical Image Analysis with Convolutional Autoencoder Neural Network. IEEE Trans. Big Data 2017, 7, 750–758. [Google Scholar] [CrossRef]
- Odena, A.; Dumoulin, V.; Olah, C. Deconvolution and Checkerboard Artifacts. Distill 2016. [Google Scholar] [CrossRef]
- Gower, J.C. A General Coefficient of Similarity and Some of Its Properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Petchey, O.L.; Gaston, K.J. Dendrograms and measures of functional diversity: A second instalment. Oikos 2009, 118, 1118–1120. [Google Scholar] [CrossRef]
- Montanari, A.; Mignani, S. Notes on the bias of dissimilarity indices for incomplete data sets: The case of archaelogical classification. Qüestiió 1994, 18, 39–49. [Google Scholar]
- Guo, X.; Gao, L.; Liu, X.; Yin, J. Improved deep embedded clustering with local structure preservation. In Proceedings of the IJCAI 2017, Melbourne Australia, 19–25 August 2017; pp. 1753–1759. [Google Scholar]
- LeCun, Y.; Cortes, C. MNIST Handwritten Digit Database. 1998. Available online: http://yann.lecun.com/exdb/mnist (accessed on 1 October 2021).
- Lewis, D.D.; Yang, Y.; Rose, T.G.; Li, F. RCV1: A New Benchmark Collection for Text Categorization Research. J. Mach. Learn. Res. 2004, 5, 361–397. [Google Scholar]
- Hadsell, R.; Chopra, S.; LeCun, Y. Dimensionality Reduction by Learning an Invariant Mapping. In Proceedings of the 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’06), New York, NY, USA, 17–22 June 2006; pp. 1735–1742. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Wallach, H., Larochelle, H., Beygelzimer, A., d’Alche Buc, F., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Van Der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Guo, X.; Liu, X.; Zhu, E.; Yin, J. Deep Clustering with Convolutional Autoencoders. In Neural Information Processing; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; pp. 373–382. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, A.; Hirschberg, J. V-Measure: A Conditional Entropy-Based External Cluster Evaluation Measure. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL), Prague, Czech Republic, 28–30 June 2007; Association for Computational Linguistics: Prague, Czech Republic, 2007; pp. 410–420. [Google Scholar]
- Das, P.; Neelima, A. A Robust Feature Descriptor for Biomedical Image Retrieval. IRBM 2020, 42, 245–257. [Google Scholar] [CrossRef]
- Camlica, Z.; Tizhoosh, H.R.; Khalvati, F. Medical image classification via SVM using LBP features from saliency-based folded data. In Proceedings of the 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 9–11 December 2015. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Advances in Neural Information Processing Systems; Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27, pp. 2672–2680. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. In Proceedings of the 2nd International Conference on Learning Representations, ICLR 2014, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]




| Number of Clusters | Train NMI AR | Train HS AR | Train NMI Mod | Train HS Mod | Test NMI AR | Test HS AR | Test NMI Mod | Test HS Mod |
|---|---|---|---|---|---|---|---|---|
| 5 | 0.397 | 0.317 | 0.743 | 0.702 | 0.386 | 0.308 | 0.744 | 0.703 |
| 10 | 0.518 | 0.516 | 0.826 | 0.887 | 0.516 | 0.504 | 0.828 | 0.890 |
| 15 | 0.525 | 0.516 | 0.811 | 0.910 | 0.529 | 0.510 | 0.805 | 0.904 |
| 20 | 0.545 | 0.536 | 0.797 | 0.898 | 0.541 | 0.533 | 0.792 | 0.898 |
| 25 | 0.565 | 0.546 | 0.793 | 0.911 | 0.584 | 0.587 | 0.793 | 0.911 |
| 30 | 0.511 | 0.514 | 0.782 | 0.897 | 0.505 | 0.508 | 0.778 | 0.892 |
| 35 | 0.544 | 0.537 | 0.754 | 0.914 | 0.528 | 0.527 | 0.752 | 0.913 |
| Silhouette Score | Train NMI AR | Train HS AR | Train NMI Mod | Train HS Mod | Test NMI AR | Test HS AR | Test NMI Mod | Test HS Mod | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.726 | 0.487 | 0.533 | 0.636 | 0.823 | 0.473 | 0.544 | 0.637 | 0.755 |
| 0.1 | 0.715 | 0.496 | 0.545 | 0.656 | 0.834 | 0.479 | 0.525 | 0.657 | 0.843 |
| 1 | 0.650 | 0.516 | 0.554 | 0.679 | 0.867 | 0.501 | 0.539 | 0.674 | 0.861 |
| 10 | 0.638 | 0.586 | 0.563 | 0.799 | 0.912 | 0.584 | 0.587 | 0.793 | 0.911 |
| 100 | 0.350 | 0.543 | 0.545 | 0.806 | 0.917 | 0.536 | 0.531 | 0.801 | 0.913 |
| Feature Descriptor | Algorithm | Test NMI AR | Test HS AR | Test NMI Modality | Test HS Modality |
|---|---|---|---|---|---|
| PCA | K-means | 0.394 | 0.342 | 0.482 | 0.633 |
| COP K-means | 0.405 | 0.473 | 0.496 | 0.643 | |
| PC K-means | 0.406 | 0.486 | 0.496 | 0.645 | |
| CAE | K-means | 0.441 | 0.523 | 0.566 | 0.745 |
| COP K-means | 0.463 | 0.545 | 0.581 | 0.771 | |
| PC K-means | 0.449 | 0.541 | 0.576 | 0.773 | |
| HOG | K-means | 0.394 | 0.451 | 0.526 | 0.659 |
| COP K-means | 0.433 | 0.452 | 0.561 | 0.677 | |
| PC K-means | 0.409 | 0.464 | 0.534 | 0.673 | |
| LBP | K-means | 0.289 | 0.291 | 0.369 | 0.478 |
| COP K-means | 0.293 | 0.351 | 0.374 | 0.490 | |
| PC K-means | 0.299 | 0.356 | 0.371 | 0.491 | |
| Convolutional IDEC | 0.473 | 0.544 | 0.637 | 0.755 | |
| Proposed model | 0.584 | 0.587 | 0.793 | 0.911 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manojlović, T.; Štajduhar, I. Deep Semi-Supervised Algorithm for Learning Cluster-Oriented Representations of Medical Images Using Partially Observable DICOM Tags and Images. Diagnostics 2021, 11, 1920. https://doi.org/10.3390/diagnostics11101920
Manojlović T, Štajduhar I. Deep Semi-Supervised Algorithm for Learning Cluster-Oriented Representations of Medical Images Using Partially Observable DICOM Tags and Images. Diagnostics. 2021; 11(10):1920. https://doi.org/10.3390/diagnostics11101920
Chicago/Turabian StyleManojlović, Teo, and Ivan Štajduhar. 2021. "Deep Semi-Supervised Algorithm for Learning Cluster-Oriented Representations of Medical Images Using Partially Observable DICOM Tags and Images" Diagnostics 11, no. 10: 1920. https://doi.org/10.3390/diagnostics11101920
APA StyleManojlović, T., & Štajduhar, I. (2021). Deep Semi-Supervised Algorithm for Learning Cluster-Oriented Representations of Medical Images Using Partially Observable DICOM Tags and Images. Diagnostics, 11(10), 1920. https://doi.org/10.3390/diagnostics11101920







