The Feasibility of Differentiating Lewy Body Dementia and Alzheimer’s Disease by Deep Learning Using ECD SPECT Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
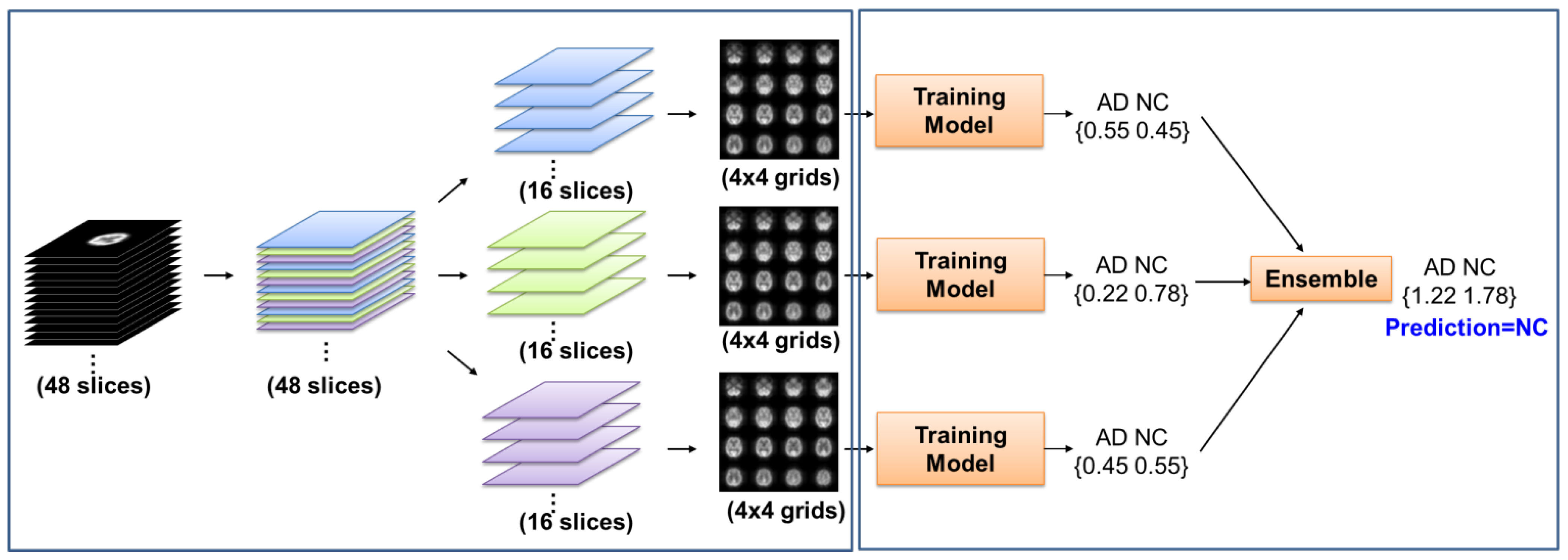
2.2. Image Acquisition and Processing
2.3. Pretrained and Training Model
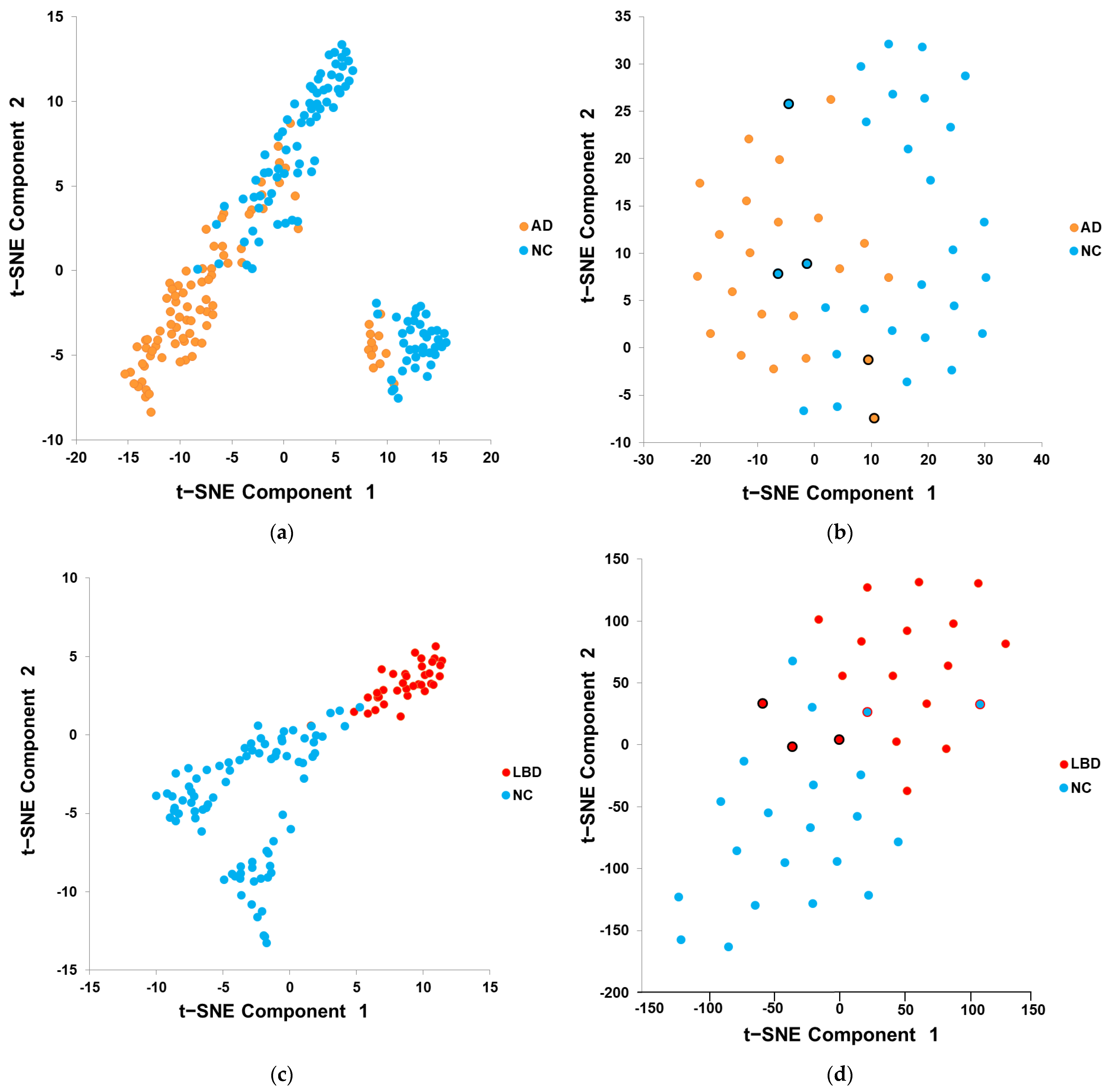
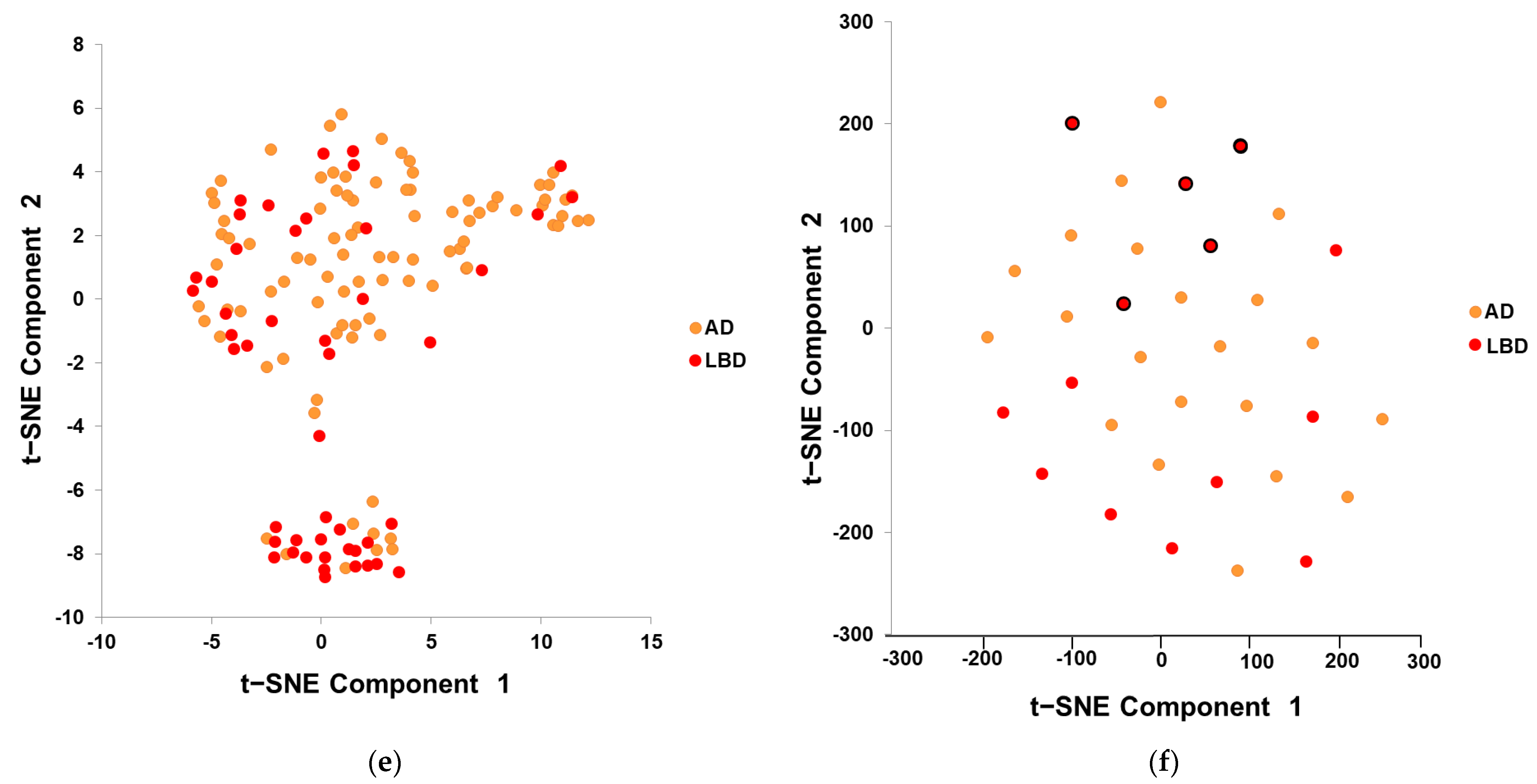
2.4. Features Visualization
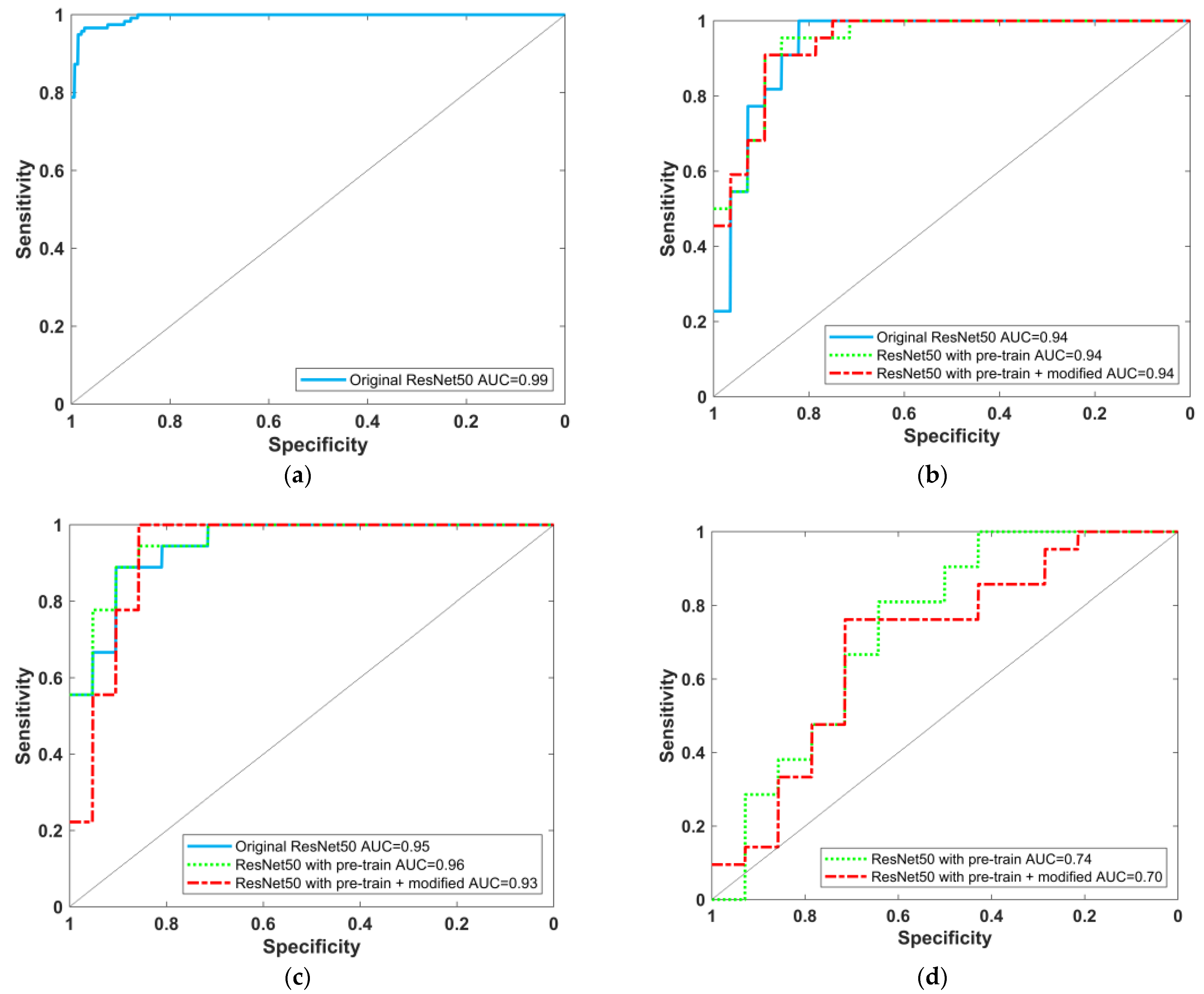
2.5. Model Testing and Evaluation
3. Results
3.1. Features Visualization
3.2. Model Testing and Result Evaluation
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taiwan Alzheimer Disease Association. Handbook of Dementia Diagnosis and Treatment; Ministry of Health and Welfare: Taipei City, Taiwan, 2017; p. 16.
- Zaccai, J.; McCracken, C.; Brayne, C. A systematic review of prevalence and incidence studies of dementia with Lewy bodies. Age Ageing 2005, 34, 561–566. [Google Scholar] [CrossRef]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- McKeith, I.G.; Boeve, B.F.; Dickson, D.W.; Halliday, G.; Taylor, J.P.; Weintraub, D.; Aarsland, D.; Galvin, J.; Attems, J.; Ballard, C.G.; et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology 2017, 89, 88–100. [Google Scholar] [CrossRef]
- Shimizu, S.; Hanyu, H.; Kanetaka, H.; Iwamoto, T.; Koizumi, K.; Abe, K. Differentiation of dementia with Lewy bodies from Alzheimer’s disease using brain SPECT. Dement. Geriatr. Cogn. Disord. 2005, 20, 25–30. [Google Scholar] [CrossRef]
- O’Brien, J.T.; Firbank, M.J.; Davison, C.; Barnett, N.; Bamford, C.; Donaldson, C.; Olsen, K.; Herholz, K.; Williams, D.; Lloyd, J. 18F-FDG PET and perfusion SPECT in the diagnosis of Alzheimer and Lewy body dementias. J. Nucl. Med. 2014, 55, 1959–1965. [Google Scholar] [CrossRef]
- Valotassiou, V.; Sifakis, N.; Papatriantafyllou, J.; Angelidis, G.; Georgoulias, P. The clinical use of SPECT and PET molecular imaging in Alzheimer’s disease. In The Clinical Spectrum of Alzheimer’s Disease—The Charge Toward Comprehensive Diagnostic and Therapeutic Strategies; De La Monte, S., Ed.; IntechOpen: London, UK, 2011; Chapter 9. [Google Scholar]
- Huang, S.H. Introduction of nuclear medicine brain scan. Chang. Gung Med. News 2017, 38, 354–355. [Google Scholar]
- Yeo, J.M.; Lim, X.X.; Khan, Z.; Pal, S. Systematic review of the diagnostic utility of SPECT imaging in dementia. Eur. Arch. Psychiatry Clin. Neurosci. 2013, 263, 539–552. [Google Scholar] [CrossRef]
- Durand-Martel, P.; Tremblay, D.; Brodeur, C.; Paquet, N. Autopsy as gold standard in FDG-PET studies in dementia. Can. J. Neurol. Sci. 2010, 37, 336–342. [Google Scholar] [CrossRef][Green Version]
- Lim, S.M.; Katsifis, A.; Villemagne, V.L.; Best, R.; Jones, G.; Saling, M.; Bradshaw, J.; Merory, J.; Woodward, M.; Hopwood, M. The 18F-FDG PET cingulate island sign and comparison to 123I-β-CIT SPECT for diagnosis of dementia with Lewy bodies. J. Nucl. Med. 2009, 50, 1638–1645. [Google Scholar] [CrossRef]
- Graff-Radford, J.; Murray, M.E.; Lowe, V.J.; Boeve, B.F.; Ferman, T.J.; Przybelski, S.A.; Lesnick, T.G.; Senjem, M.L.; Gunter, J.L.; Smith, G.E. Dementia with Lewy bodies Basis of cingulate island sign. Neurology 2014, 83, 801–809. [Google Scholar] [CrossRef]
- Iizuka, T.; Kameyama, M. Cingulate island sign on FDG-PET is associated with medial temporal lobe atrophy in dementia with Lewy bodies. Ann. Nucl. Med. 2016, 30, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Imabayashi, E.; Soma, T.; Sone, D.; Tsukamoto, T.; Kimura, Y.; Sato, N.; Murata, M.; Matsuda, H. Validation of the cingulate island sign with optimized ratios for discriminating dementia with Lewy bodies from Alzheimer’s disease using brain perfusion SPECT. Ann. Nucl. Med. 2017, 31, 536–543. [Google Scholar] [CrossRef]
- Iizuka, T.; Fukasawa, M.; Kameyama, M. Deep-learning-based imaging-classification identified cingulate island sign in dementia with Lewy bodies. Sci. Rep. 2019, 9, 1–9. [Google Scholar]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Valliani, A.A.; Ranti, D.; Oermann, E.K. Deep Learning and Neurology: A Systematic Review. Neurol. Ther. 2019, 8, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Nensa, F.; Demircioglu, A.; Rischpler, C. Artificial Intelligence in Nuclear Medicine. J. Nucl. Med. 2019, 60 (Suppl. S2), 29S–37S. [Google Scholar] [CrossRef]
- Pellegrini, E.; Ballerini, L.; Hernandez, M.D.C.V.; Chappell, F.M.; González-Castro, V.; Anblagan, D.; Danso, S.; Muñoz-Maniega, S.; Job, D.; Pernet, C.; et al. Machine learning of neuroimaging for assisted diagnosis of cognitive impairment and dementia: A systematic review. Alzheimers Dement 2018, 11, 519–535. [Google Scholar] [CrossRef]
- Lopez-Martin, M.; Nevado, A.; Carro, B. Detection of early stages of Alzheimer’s disease based on MEG activity with a randomized convolutional neural network. Artif. Intell. Med. 2020, 107, 101924. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Sohn, J.H.; Kawczynski, M.G.; Trivedi, H.; Harnish, R.; Jenkins, N.W.; Lituiev, D.; Copeland, T.P.; Aboian, M.S.; Mari Aparici, C. A Deep Learning Model to Predict a Diagnosis of Alzheimer Disease by Using 18F-FDG PET of the Brain. Radiology 2019, 290, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Elazab, A.; Yang, P.; Wang, T.; Zhou, F.; Hu, H.; Xiao, X.; Lei, B. Deep Learning Framework for Alzheimer’s Disease Diagnosis via 3D-CNN and FSBi-LSTM. IEEE Access 2019, 7, 63605–63618. [Google Scholar] [CrossRef]
- Choi, H.; Kim, Y.K.; Yoon, E.J.; Lee, J.Y.; Lee, D.S. Cognitive signature of brain FDG PET based on deep learning: Domain transfer from Alzheimer’s disease to Parkinson’s disease. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 403–412. [Google Scholar] [CrossRef]
- Ni, Y.C.; Tseng, F.P.; Pai, M.C.; Hsiao, T.; Lin, K.J.; Lin, Z.K.; Lin, W.B.; Chiu, P.Y.; Hung, G.U.; Chang, C.C.; et al. Detection of Alzheimer’s disease using ECD SPECT images by transfer learning from FDG PET. Ann. Nucl. Med. 2021, 35, 889–899. [Google Scholar] [CrossRef]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, preprint. arXiv:1412.6980. [Google Scholar]
- van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn Res. 2008, 9, 2579–2605. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn Res. 2011, 12, 2825–2830. [Google Scholar]
- Segovia, F.; García-Pérez, M.; Górriz, J.M.; Ramírez, J.; Martínez-Murcia, F.J. Assisting the diagnosis of neurodegenerative disorders using principal component analysis and TensorFlow. In International Joint Conference SOCO’16-CISIS’16-ICEUTE’16; Graña, M., López-Guede, J.M., Etxaniz, O., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 43–52. [Google Scholar]
- Świetlik, D.; Białowąs, J. Application of Artificial Neural Networks to Identify Alzheimer’s Disease Using Cerebral Perfusion SPECT Data. Int. J. Environ. Res. Public Health 2019, 16, 1303. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]

| Characteristic | NC | AD | LBD |
|---|---|---|---|
| Number of subjects | 134 | 113 | 61 |
| Age at the time of SPECT (years) | 67.0 ± 8.5 | 74.4 ± 7.0 | 77.2 ± 5.9 |
| Sex (F:M) | 88:46 | 58:55 | 25:36 |
| MMSE | 27.5 ± 2.4 | 19.2 ± 5.3 | 17.6 ± 5.9 |
| CDR | 0.22 ± 0.25 | 0.79 ± 0.39 | 0.93 ± 0.50 |
| Characteristic | NC | AD |
|---|---|---|
| Number of subjects | 666 | 667 |
| Age at the time of SPECT (years) | 76.4 ± 5.7 | 76.8 ± 7.5 |
| Sex (F:M) | 282:384 | 268:399 |
| MMSE | 28.5 ± 4.0 | 21.9 ± 5.1 |
| CDR | 0.03 ± 0.16 | 0.83 ± 0.41 |
| Task | Training Data Set (80%) | Testing Data Set (20%) |
|---|---|---|
| ADNI Pretrained AD/NC | 549/517 (total: 1066) | 118/149 (total: 267) |
| AD/NC | 91/106 (total: 197) | 22/28 (total: 50) |
| LBD/NC | 43/113 (total: 156) | 18/21 (total: 39) |
| AD/LBD | 92/47 (total: 139) | 21/14 (total: 35) |
| Method | Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1 Score (%) | AUC for AD/NC (95% CI) | |
|---|---|---|---|---|---|---|---|
| Proposed (ECD image) | Original ResNet-50 model | 90.91 (20/22) | 50.00 (14/28) | 58.82 (20/34) | 68.00 (34/50) | 71.43 | 0.94 (0.82–0.99) |
| ResNet-50 model (with ADNI pretrain) | 95.45 (21/22) | 78.57 (22/28) | 77.78 (21/27) | 86.00 (43/50) | 85.71 | 0.94 (0.86~0.99) | |
| ResNet-50 model (with ADNI pretrain + modified) | 90.91 (20/22) | 89.29 (25/28) | 86.96 (20/23) | 90.00 (45/50) | 88.89 | 0.94 (0.84–0.98) | |
| Reference (ECD image) | 3 layers DNN 1,+ | 95.12 | 75.00 | - | 83.51 | - | - |
| Naive Bayes + | 68.29 | 91.07 | - | 81.44 | - | - | |
| Decision trees + | 78.05 | 85.71 | - | 82.47 | - | - | |
| SVM + | 82.92 | 82.14 | - | 82.47 | - | - | |
| Reference (nonECD image) | CNN 2,* (I-123-IMP 3D-SSP) | - | - | - | 92.39 | - | 0.94 |
| ANN 3,^ (HMPAO 36 value) | 93.80 | 100.00 | - | - | - | 0.97 |
| Method | Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1 Score (%) | AUC for LBD/NC (95% CI) | |
|---|---|---|---|---|---|---|---|
| Proposed (ECD image) | Original ResNet-50 model | 83.33 (15/18) | 90.48 (19/21) | 88.24 (15/17) | 87.18 (34/39) | 85.71 | 0.95 (0.83–0.99) |
| ResNet-50 model (with ADNI pretrain) | 94.44 (17/18) | 76.19 (16/21) | 77.27 (17/22) | 84.62 (33/39) | 84.99 | 0.96 (0.83–0.99) | |
| ResNet-50 model(with ADNI pretrain + modified) | 100.00 (18/18) | 71.43 (15/21) | 75.00 (18/24) | 84.62 (33/39) | 85.71 | 0.93 (0.78–0.99) | |
| Reference (nonECD image) | CNN * (I-123-IMP 3D-SSP) | - | - | - | 93.07 | - | 0.95 |
| Method | Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1 Score (%) | AUC for AD/LBD (95% CI) | |
|---|---|---|---|---|---|---|---|
| Proposed (ECD image) | Original ResNet-50 model | Training unsuccessful | |||||
| ResNet-50 model (with ADNI pretrain) | 76.19 (16/21) | 64.29 (9/14) | 76.19 (16/21) | 71.43 (25/35) | 76.19 | 0.74 (0.52–0.90) | |
| ResNet-50 model(with ADNI pretrain + modified) | 76.19 (16/21) | 57.14 (8/14) | 72.73 (16/22) | 68.57 (24/35) | 74.42 | 0.70 (0.47–0.86) | |
| Reference (nonECD image) | CNN * (I-123-IMP 3D-SSP) | - | - | - | 89.32 | - | 0.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Y.-C.; Tseng, F.-P.; Pai, M.-C.; Hsiao, I.-T.; Lin, K.-J.; Lin, Z.-K.; Lin, C.-Y.; Chiu, P.-Y.; Hung, G.-U.; Chang, C.-C.; et al. The Feasibility of Differentiating Lewy Body Dementia and Alzheimer’s Disease by Deep Learning Using ECD SPECT Images. Diagnostics 2021, 11, 2091. https://doi.org/10.3390/diagnostics11112091
Ni Y-C, Tseng F-P, Pai M-C, Hsiao I-T, Lin K-J, Lin Z-K, Lin C-Y, Chiu P-Y, Hung G-U, Chang C-C, et al. The Feasibility of Differentiating Lewy Body Dementia and Alzheimer’s Disease by Deep Learning Using ECD SPECT Images. Diagnostics. 2021; 11(11):2091. https://doi.org/10.3390/diagnostics11112091
Chicago/Turabian StyleNi, Yu-Ching, Fan-Pin Tseng, Ming-Chyi Pai, Ing-Tsung Hsiao, Kun-Ju Lin, Zhi-Kun Lin, Chia-Yu Lin, Pai-Yi Chiu, Guang-Uei Hung, Chiung-Chih Chang, and et al. 2021. "The Feasibility of Differentiating Lewy Body Dementia and Alzheimer’s Disease by Deep Learning Using ECD SPECT Images" Diagnostics 11, no. 11: 2091. https://doi.org/10.3390/diagnostics11112091
APA StyleNi, Y.-C., Tseng, F.-P., Pai, M.-C., Hsiao, I.-T., Lin, K.-J., Lin, Z.-K., Lin, C.-Y., Chiu, P.-Y., Hung, G.-U., Chang, C.-C., Chang, Y.-T., Chuang, K.-S., & Alzheimer’s Disease Neuroimaging Initiative. (2021). The Feasibility of Differentiating Lewy Body Dementia and Alzheimer’s Disease by Deep Learning Using ECD SPECT Images. Diagnostics, 11(11), 2091. https://doi.org/10.3390/diagnostics11112091






