Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm
Abstract
:1. Introduction
2. Liquid Biopsy
3. Clinical Applications of ctDNA
4. Early Cancer Diagnosis
5. Empirical Calculations Challenging the Clinical Utility of Grail and Similar Tests for Early Cancer Diagnosis
6. Additional challenges with Grail and Related Technologies
7. Experimental Data from Grail That Support our Predictions
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lo, Y.D. Screening of Fetal Chromosomal Aneuploidy by Noninvasive Prenatal Testing: From Innovation to Setting Public Health Agendas to Potential Impact on Other Fields. Clin. Chem. 2019, 66, 25–28. [Google Scholar] [CrossRef]
- Vermeesch, J.R.; Voet, T.; Devriendt, K. Prenatal and pre-implantation genetic diagnosis. Nat. Rev. Genet. 2016, 17, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Asante, D.-B.; Calapre, L.; Ziman, M.; Meniawy, T.M.; Gray, E.S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020, 468, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nat. Cell Biol. 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heitzer, E.; Speicher, M.R. One size does not fit all: Size-based plasma DNA diagnostics. Sci. Transl. Med. 2018, 10, eaav3873. [Google Scholar] [CrossRef]
- Bredno, J.; Lipson, J.; Venn, O.; Aravanis, A.M.; Jamshidi, A. Clinical correlates of circulating cell-free DNA tumor fraction. PLoS ONE 2021, 16, e0256436. [Google Scholar] [CrossRef]
- Tan, G.; Chu, C.; Gui, X.; Li, J.; Chen, Q. The prognostic value of circulating cell-free DNA in breast cancer. Med. 2018, 97, e0197. [Google Scholar] [CrossRef]
- Stover, D.; Parsons, H.A.; Ha, G.; Freeman, S.; Barry, W.T.; Guo, H.; Choudhury, A.; Gydush, G.; Reed, S.; Rhoades, J.; et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 543–553. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chang, C.-W.; Spoerke, J.M.; Yoh, K.E.; Kapoor, V.; Baudo, C.D.; Aimi, J.; Yu, M.; Liang-Chu, M.M.; Suttmann, R.; et al. Low-pass Whole-genome Sequencing of Circulating Cell-free DNA Demonstrates Dynamic Changes in Genomic Copy Number in a Squamous Lung Cancer Clinical Cohort. Clin. Cancer Res. 2019, 25, 2254–2263. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhang, Y.; Cheng, Y.; Zhang, D.; Zhu, S.; Ma, X. Prognostic value of circulating cell-free DNA in patients with pancreatic cancer: A systemic review and meta-analysis. Gene 2018, 679, 328–334. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, 2415. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Dong, Z.; Hubbell, E.; Kurtzman, K.N.; Oxnard, G.R.; Venn, O.; Melton, C.; Clarke, C.A.; Shaknovich, R.; Ma, T.; et al. Prognostic Significance of Blood-Based Multi-cancer Detection in Plasma Cell-Free DNA. Clin. Cancer Res. 2021, 27, 4221–4229. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Saw, R.; Thompson, J.; Lo, S.; Spillane, A.; Shannon, K.; Stretch, J.; Howle, J.; Menzies, A.; Carlino, M.; et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 2019, 30, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Balgkouranidou, I.; Chimonidou, M.; Milaki, G.; Tsarouxa, E.G.; Kakolyris, S.; Welch, D.R.; Georgoulias, V.; Lianidou, E.S. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br. J. Cancer 2014, 110, 2054–2062. [Google Scholar] [CrossRef] [Green Version]
- Fiala, C.; Diamandis, E.P. Mutations in normal tissues—Some diagnostic and clinical implications. BMC Med. 2020, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Strijker, M.; Soer, E.C.; De Pastena, M.; Creemers, A.; Balduzzi, A.; Beagan, J.; Busch, O.R.; Van Delden, O.M.; Halfwerk, H.; Van Hooft, J.E.; et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int. J. Cancer 2019, 146, 1445–1456. [Google Scholar] [CrossRef]
- Lee, J.H.; Menzies, A.M.; Carlino, M.S.; McEvoy, A.C.; Sandhu, S.; Weppler, A.M.; Diefenbach, R.J.; Dawson, S.-J.; Kefford, R.; Millward, M.J.; et al. Longitudinal Monitoring of ctDNA in Patients with Melanoma and Brain Metastases Treated with Immune Checkpoint Inhibitors. Clin. Cancer Res. 2020, 26, 4064–4071. [Google Scholar] [CrossRef] [Green Version]
- Openshaw, M.; Page, K.; Garcia, D.F.; Guttery, D.; Shaw, J.A. The role of ctDNA detection and the potential of the liquid biopsy for breast cancer monitoring. Expert Rev. Mol. Diagn. 2016, 16, 751–755. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Fiala, C.; Diamandis, E.P. Can a Broad Molecular Screen Based on Circulating Tumor DNA Aid in Early Cancer Detection? J. Appl. Lab. Med. 2020, 5, 1372–1377. [Google Scholar] [CrossRef]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Oxnard, G.; Klein, E.; Swanton, C.; Seiden, M.; Smith, D.; Richards, D.; Yeatman, T.J.; Cohn, A.L.; Lapham, R.; et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; El Messaoudi, S.; Pang, D.; Dritschilo, A.; Thierry, A.R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 2014, 8, 927–941. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Peyrotte, E.A.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nat. Cell Biol. 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Duffy, M.J.; Diamandis, E.P.; Crown, J. Circulating tumor DNA (ctDNA) as a pan-cancer screening test: Is it finally on the horizon? Clin. Chem. Lab. Med. 2021, 59, 1353–1361. [Google Scholar] [CrossRef]
- Fiala, C.; Diamandis, E.P. Can Grail find the trail to early cancer detection? Clin. Chem. Lab. Med. 2019, 57, 403–406. [Google Scholar] [CrossRef]
- Diamandis, E.P.; Fiala, C. Can circulating tumor DNA be used for direct and early stage cancer detection? F1000Research 2017, 6, 2129. [Google Scholar] [CrossRef] [Green Version]
- Fiala, C.; Diamandis, E.P. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 2018, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; The National Lung Screening Trial Research Team; et al. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [Green Version]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Del Monte, U. Does the cell number 109still really fit one gram of tumor tissue? Cell Cycle 2009, 8, 505–506. [Google Scholar] [CrossRef] [Green Version]
- Narod, S. Disappearing Breast Cancers. Curr. Oncol. 2012, 19, 59–60. [Google Scholar] [CrossRef] [Green Version]
- Weedon-Fekjaer, H.; Lindqvist, B.H.; Vatten, L.J.; Aalen, O.O.; Tretli, S. Breast cancer tumor growth estimated through mammography screening data. Breast Cancer Res. 2008, 10, R41. [Google Scholar] [CrossRef] [Green Version]
- Diamandis, E.; Li, M. The side effects of translational omics: Overtesting, overdiagnosis, overtreatment. Clin. Chem. Lab. Med. 2015, 54, 389–396. [Google Scholar] [CrossRef]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Jones, P.H.; Wedge, D.; Sale, J.; Campbell, P.J.; Nik-Zainal, S.; Stratton, M.R. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015, 47, 1402–1407. [Google Scholar] [CrossRef] [Green Version]
- Schwaederle, M.; Husain, H.; Fanta, P.T.; Piccioni, D.; Kesari, S.; Schwab, R.B.; Banks, K.; Lanman, R.B.; Talasaz, A.; Parker, B.A.; et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget 2016, 7, 9707–9717. [Google Scholar] [CrossRef]
- Gormally, E.; Vineis, P.; Matullo, G.; Veglia, F.; Caboux, E.; Le Roux, E.; Peluso, M.; Garte, S.; Guarrera, S.; Munnia, A.; et al. TP53 and KRAS2 Mutations in Plasma DNA of Healthy Subjects and Subsequent Cancer Occurrence: A Prospective Study. Cancer Res. 2006, 66, 6871–6876. [Google Scholar] [CrossRef] [Green Version]
- Diamandis, E. Cancer dynamics and the success of cancer screening programs. Clin. Chem. Lab. Med. 2016, 54, e211–e212. [Google Scholar] [CrossRef]
- Liu, M.C.; Maddala, T.; Aravanis, A.; Hubbell, E.; Beausang, J.F.; Filippova, D.; Gross, S.; Jamshidi, A.; Kurtzman, K.; Shen, L.; et al. Breast cancer cell-free DNA (cfDNA) profiles reflect underlying tumor biology: The Circulating Cell-Free Genome Atlas (CCGA) study. J. Clin. Oncol. 2018, 36, 536. [Google Scholar] [CrossRef] [Green Version]


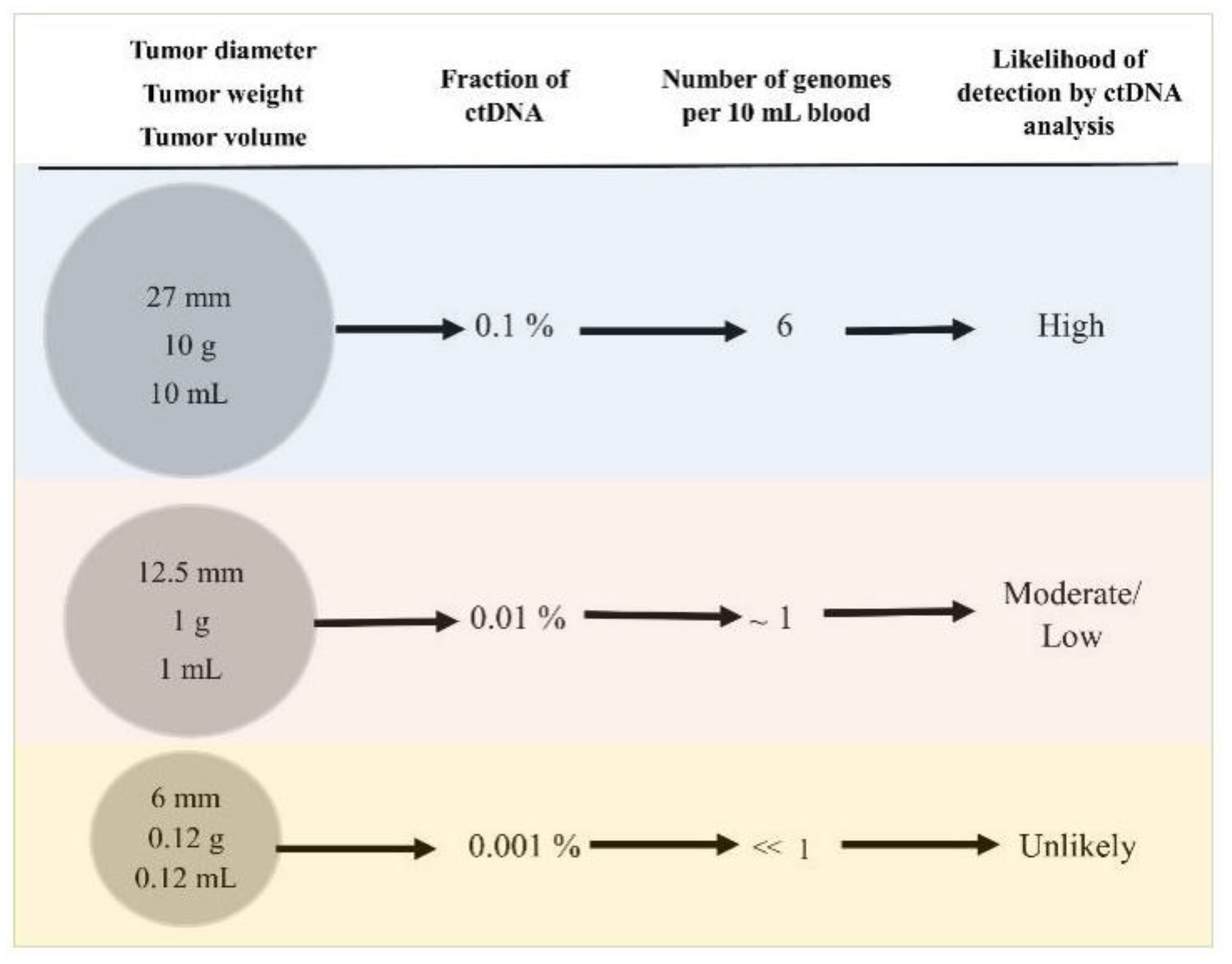
| Tumor Diameter, mm | Tumor Weight, mg | Tumor Volumen mL (cm3) | Number of Cancer Cells | Percentage Fraction of Mutant ctDNA | Number of Cancer Genomes per 10 mL of Blood | Chance of Progression c | Mammographic Screen Sensitivity d |
|---|---|---|---|---|---|---|---|
| 27 | 10,000 | 10 a | 10,000,000,000 | 1:1000 | 6 | - | - |
| 12.5 | 1000 | 1 b | 1,000,000,000 | 1:10,000 | 0.6 | - | - |
| 10 | 500 | 0.5 | 500,000,000 | 1:20,000 | 0.3 | 50% | 91% |
| 8 | 250 | 0.25 | 250,000,000 | 1:40,000 | 0.15 | 25% | - |
| 6 | 125 | 0.12 | 125,000,000 | 1:80,000 | <0.1 | - | - |
| 5 | 62 | 0.06 | 62,000,000 | 1:160,000 | <0.1 | 6% | 26% |
| 4 | 31 | 0.03 | 32,000,000 | 1:320,000 | <0.1 | - | - |
| 3 | 16 | 0.015 | 16,000,000 | 1:640,000 | <0.1 | - | - |
| 2.4 | 8 | 0.007 | 8,000,000 | 1:1,300,000 | <0.1 | - | - |
| 2 | 4 | 0.0035 | 4,000,000 | 1:2,600,000 | <0.1 | - | - |
| 1.5 | 2 | 0.0017 | 2,000,000 | 1:5,200,000 | <0.1 | - | - |
| 1.1 | 1 | 0.0008 | 1,000,000 | 1:10,000,000 | <0.1 | 0.05% | - |
| Benefits | Harms |
|---|---|
| Identification of disease predisposition or early diagnosis, leading to prevention or effective therapy. | If no treatment or prevention available, diagnosis may cause anxiety/depression. False-positives leading to more testing; some testing may be invasive or have side effects (biopsies, surgeries, anxiety, depression). Incidental findings/indolent disease 1 (over-diagnosis, over-treatment, and some treatments may be invasive, have serious side effects, and be costly). Harms of testing (e.g., radiation, bleeding, colon perforation). Cost-effectiveness. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pons-Belda, O.D.; Fernandez-Uriarte, A.; Diamandis, E.P. Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm. Diagnostics 2021, 11, 2171. https://doi.org/10.3390/diagnostics11122171
Pons-Belda OD, Fernandez-Uriarte A, Diamandis EP. Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm. Diagnostics. 2021; 11(12):2171. https://doi.org/10.3390/diagnostics11122171
Chicago/Turabian StylePons-Belda, Oscar D., Amaia Fernandez-Uriarte, and Eleftherios P. Diamandis. 2021. "Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm" Diagnostics 11, no. 12: 2171. https://doi.org/10.3390/diagnostics11122171
APA StylePons-Belda, O. D., Fernandez-Uriarte, A., & Diamandis, E. P. (2021). Can Circulating Tumor DNA Support a Successful Screening Test for Early Cancer Detection? The Grail Paradigm. Diagnostics, 11(12), 2171. https://doi.org/10.3390/diagnostics11122171





