Estimating Glomerular Filtration Rate from Serum Myo-Inositol, Valine, Creatinine and Cystatin C
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Laboratory Methods
2.2.1. mGFR, Serum Creatinine and Cystatin C Measurements
2.2.2. eGFR
2.2.3. NMR Analysis and Biomarker Quantification
2.3. Biostatistical Methods
2.3.1. Metrics for Performance Evaluation and Benchmarking
2.3.2. Development of Equations and Model Training
2.3.3. Selection, Model Engineering and Internal Validation of Equations
2.3.4. External Validation of the New Equation
3. Results
3.1. Characteristics of Participants
3.2. Formulation and Selection of Candidate Equations
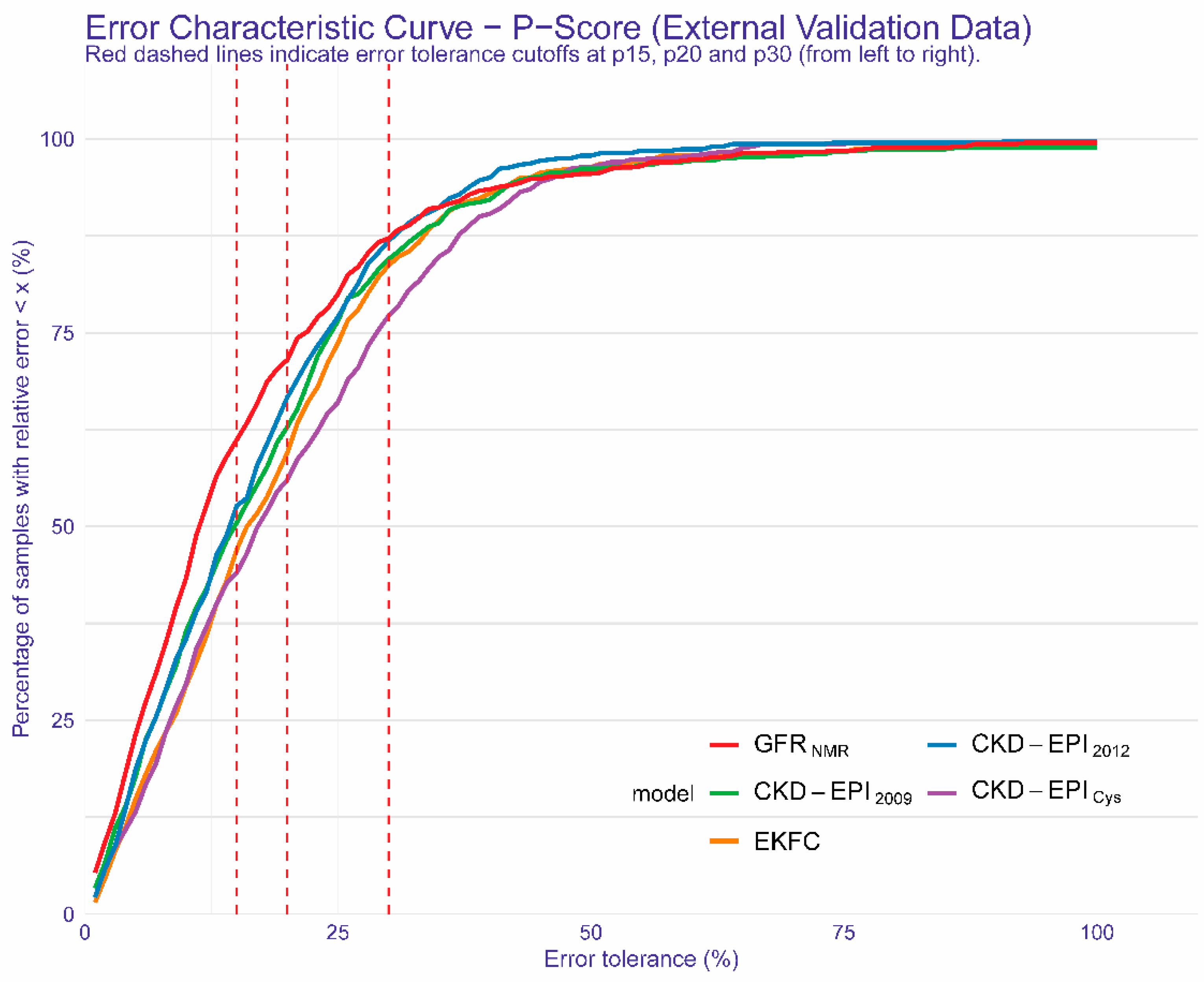
3.3. The New GFRNMR Equation and Its Performance in the External Validation Dataset
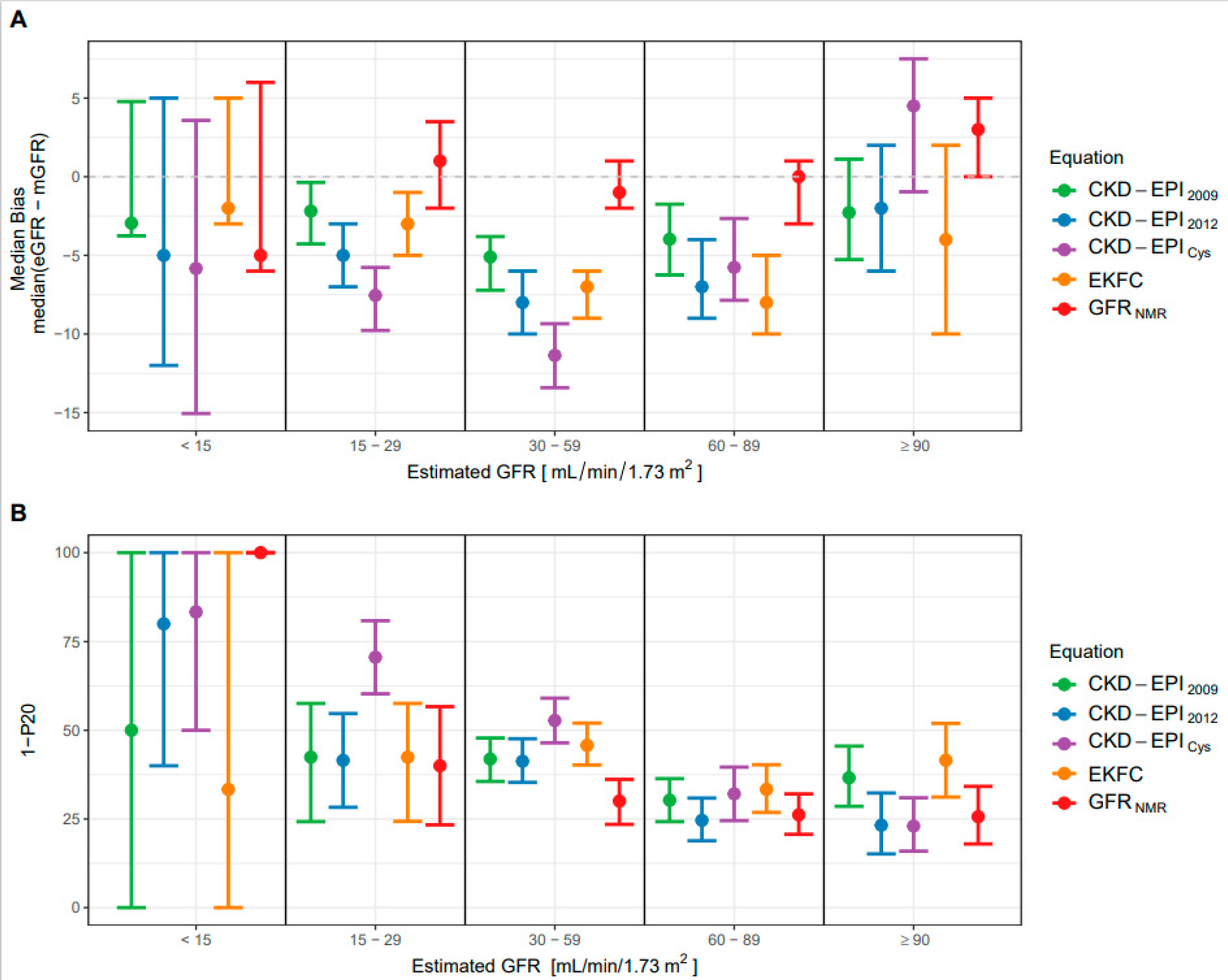
3.4. Performance of the New GFRNMR Equation in Subpopulations of the External Validation Dataset
3.5. Evaluation of the Potential Clinical Benefit of the New GFRNMR Equation
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F.; Feldman, H.I.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Greene, T.; et al. A New Equation to Estimate Glomerular Filtration Rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef]
- Inker, L.A.; Schmid, C.H.; Tighiouart, H.; Eckfeldt, J.H.; Feldman, H.I.; Greene, T.; Kusek, J.W.; Manzi, J.; Van Lente, F.; Zhang, Y.L.; et al. Estimating Glomerular Filtration Rate from Serum Creatinine and Cystatin C. N. Engl. J. Med. 2012, 367, 20–29. [Google Scholar] [CrossRef]
- Pottel, H.; Björk, J.; Courbebaisse, M.; Couzi, L.; Ebert, N.; Eriksen, B.O.; Dalton, R.N.; Dubourg, L.; Gaillard, F.; Garrouste, C.; et al. Development and Validation of a Modified Full Age Spectrum Creatinine-Based Equation to Estimate Glomerular Filtration Rate: A Cross-Sectional Analysis of Pooled Data. Ann. Intern. Med. 2021, 174, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.S.; Tighiouart, H.; Inker, L.A. Improving Glomerular Filtration Rate Estimation-Across the Age and Diversity Spectrum. Ann. Intern. Med. 2021, 174, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Bansal, N. Measured GFR as “Gold Standard”—All That Glitters Is Not Gold? Clin. J. Am. Soc. Nephrol. 2011, 6, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Porrini, E.; Ruggenenti, P.; Luis-Lima, S.; Carrara, F.; Jiménez, A.; de Vries, A.P.J.; Torres, A.; Gaspari, F.; Remuzzi, G. Estimated GFR: Time for a Critical Appraisal. Nat. Rev. Nephrol. 2019, 15, 177–190. [Google Scholar] [CrossRef]
- CKD Evaluation and Management—KDIGO. Available online: https://kdigo.org/guidelines/ckd-evaluation-and-management/ (accessed on 9 July 2021).
- KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int. Suppl. 2013, 3, 1–150.
- Steubl, D.; Inker, L.A. How Best to Estimate Glomerular Filtration Rate? Novel Filtration Markers and Their Application. Curr. Opin. Nephrol. Hypertens. 2018, 27, 398–405. [Google Scholar] [CrossRef]
- Sekula, P.; Goek, O.-N.; Quaye, L.; Barrios, C.; Levey, A.S.; Römisch-Margl, W.; Menni, C.; Yet, I.; Gieger, C.; Inker, L.A.; et al. A Metabolome-Wide Association Study of Kidney Function and Disease in the General Population. J. Am. Soc. Nephrol. 2016, 27, 1175–1188. [Google Scholar] [CrossRef] [PubMed]
- Coresh, J.; Inker, L.A.; Sang, Y.; Chen, J.; Shafi, T.; Post, W.S.; Shlipak, M.G.; Ford, L.; Goodman, K.; Perichon, R.; et al. Metabolomic Profiling to Improve Glomerular Filtration Rate Estimation: A Proof-of-Concept Study. Nephrol. Dial. Transplant. 2019, 34, 825–833. [Google Scholar] [CrossRef]
- Ehrich, J.; Dubourg, L.; Hansson, S.; Pape, L.; Steinle, T.; Fruth, J.; Höckner, S.; Schiffer, E. Serum Myo-Inositol, Dimethyl Sulfone, and Valine in Combination with Creatinine Allow Accurate Assessment of Renal Insufficiency-A Proof of Concept. Diagnostics 2021, 11, 234. [Google Scholar] [CrossRef]
- Coresh, J.; Selvin, E.; Stevens, L.A.; Manzi, J.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Levey, A.S. Prevalence of Chronic Kidney Disease in the United States. JAMA 2007, 298, 2038–2047. [Google Scholar] [CrossRef]
- Shi, X.; Shi, Y.; Zhang, L.; Gan, L.; Zhong, X.; Huang, Y.; Yao, C.; Wang, Y.; Dong, C.; Liu, B.; et al. Analysis of Chronic Kidney Disease among National Hospitalization Data with 14 Million Children. BMC Nephrol. 2021, 22, 195. [Google Scholar] [CrossRef] [PubMed]
- Schaeffner, E.S.; van der Giet, M.; Gaedeke, J.; Tölle, M.; Ebert, N.; Kuhlmann, M.K.; Martus, P. The Berlin Initiative Study: The Methodology of Exploring Kidney Function in the Elderly by Combining a Longitudinal and Cross-Sectional Approach. Eur. J. Epidemiol. 2010, 25, 203–210. [Google Scholar] [CrossRef]
- Ebert, N.; Loesment, A.; Martus, P.; Jakob, O.; Gaedeke, J.; Kuhlmann, M.; Bartel, J.; Schuchardt, M.; Tölle, M.; Huang, T.; et al. Iohexol Plasma Clearance Measurement in Older Adults with Chronic Kidney Disease—Sampling Time Matters. Nephrol. Dial. Transplant. 2015, 30, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Selistre, L.; Rech, D.L.; de Souza, V.; Iwaz, J.; Lemoine, S.; Dubourg, L. Diagnostic Performance of Creatinine-Based Equations for Estimating Glomerular Filtration Rate in Adults 65 Years and Older. JAMA Intern. Med. 2019, 179, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Grewal, G.S.; Blake, G.M. Reference Data for 51Cr-EDTA Measurements of the Glomerular Filtration Rate Derived from Live Kidney Donors. Nucl. Med. Commun. 2005, 26, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M.; Bergert, J.H.; Larson, T.S.; Liedtke, R.R. GFR Determined by Nonradiolabeled Iothalamate Using Capillary Electrophoresis. Am. J. Kidney Dis. 1997, 30, 646–652. [Google Scholar] [CrossRef]
- Soveri, I.; Berg, U.B.; Björk, J.; Elinder, C.-G.; Grubb, A.; Mejare, I.; Sterner, G.; Bäck, S.-E.; SBU GFR Review Group. Measuring GFR: A Systematic Review. Am. J. Kidney Dis. 2014, 64, 411–424. [Google Scholar] [CrossRef]
- Thienpont, L.M.; Van Landuyt, K.G.; Stöckl, D.; De Leenheer, A.P. Candidate Reference Method for Determining Serum Creatinine by Isocratic HPLC: Validation with Isotope Dilution Gas Chromatography-Mass Spectrometry and Application for Accuracy Assessment of Routine Test Kits. Clin. Chem. 1995, 41, 995–1003. [Google Scholar] [CrossRef]
- Levin, A.S.; Bilous, R.W.; Coresh, J. Chapter 1: Definition and Classification of CKD. Kidney Int. Suppl. 2013, 3, 19–62. [Google Scholar] [CrossRef]
- Aguilar, J.A.; Nilsson, M.; Bodenhausen, G.; Morris, G.A. Spin Echo NMR Spectra without J Modulation. Chem. Commun. 2011, 48, 811–813. [Google Scholar] [CrossRef]
- R Core Team. R: The R Project for Statistical Computing. R Package Version 3.5.3. 2020. Available online: https://www.r-project.org/ (accessed on 9 June 2021).
- Hunt, T.; ModelMetrics: Rapid Calculation of Model Metrics. R Package Version 1.2.2.2. 2020. Available online: https://CRAN.R-project.org/package=ModelMetrics (accessed on 26 July 2021).
- Gosiewska, A.; Biecek, P. Auditor: An R Package for Model-Agnostic Visual Validation and Diagnostics. R J. 2019, 11, 85–98. [Google Scholar] [CrossRef]
- Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P.; Short, T.; Lianoglou, S.; Antonyan, E.; Bonsch, M.; Parsonage, H.; et al. Data.Table: Extension of “Data.Frame”. R Package Version 1.13.2. 2020. Available online: https://CRAN.R-project.org/package=data.table (accessed on 26 July 2021).
- Biecek, P.; Kosiński, M. Archivist: An R Package for Managing, Recording and Restoring Data Analysis Results. J. Stat. Softw. 2017, 82, 1–28. [Google Scholar] [CrossRef]
- Davison, A.C.; Hinkley, D.V. Bootstrap Methods and Their Application; Cambridge Series in Statistical and Probabilistic Mathematics; Cambridge University Press: Cambridge, UK, 1997; ISBN 0-521-57391-2. Available online: http://statwww.epfl.ch/davison/BMA/ (accessed on 26 July 2021).
- Canty, A.; Ripley, B.D.; Boot: Bootstrap R (S-Plus) Functions. R Package Version 1.3-25. 2020. Available online: http://statwww.epfl.ch/davison/BMA/library.html (accessed on 26 July 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Siegel, S. Nonparametric Statistics for the Behavioral Sciences; McGraw-Hill: New York, NY, USA, 1956; ISBN 978-0-07-085689-9. [Google Scholar]
- McNemar, Q. Note on the Sampling Error of the Difference between Correlated Proportions or Percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef]
- Willmott, C.J.; Matsuura, K. Advantages of the Mean Absolute Error (MAE) over the Root Mean Square Error (RMSE) in Assessing Average Model Performance. Clim. Res. 2005, 30, 79–82. [Google Scholar] [CrossRef]
- Pencina, M.J.; D’Agostino, R.B.; D’Agostino, R.B.; Vasan, R.S. Evaluating the Added Predictive Ability of a New Marker: From Area under the ROC Curve to Reclassification and Beyond. Stat. Med. 2008, 27, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Dreiseitl, S.; Ohno-Machado, L.; Binder, M. Comparing Three-Class Diagnostic Tests by Three-Way ROC Analysis. Med. Decis. Mak. 2000, 20, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Nakas, C.T.; Yiannoutsos, C.T. Ordered Multiple-Class ROC Analysis with Continuous Measurements. Stat. Med. 2004, 23, 3437–3449. [Google Scholar] [CrossRef] [PubMed]
- Vuong, Q.H. Likelihood Ratio Tests for Model Selection and Non-Nested Hypotheses. Econometrica 1989, 57, 307–333. [Google Scholar] [CrossRef]
- Merkle, E.; You, D.; Schneider, L.; Bae, S.; Nonnest2: Tests of Non-Nested Models. R Package Version 0.5-5. 2020. Available online: https://CRAN.R-project.org/package=nonnest2 (accessed on 26 July 2021).
- Go, A.S.; Chertow, G.M.; Fan, D.; McCulloch, C.E.; Hsu, C. Chronic Kidney Disease and the Risks of Death, Cardiovascular Events, and Hospitalization. N. Engl. J. Med. 2004, 351, 1296–1305. [Google Scholar] [CrossRef]
- Peralta, C.A.; Shlipak, M.G.; Judd, S.; Cushman, M.; McClellan, W.; Zakai, N.A.; Safford, M.M.; Zhang, X.; Muntner, P.; Warnock, D. Detection of Chronic Kidney Disease with Creatinine, Cystatin C, and Urine Albumin-to-Creatinine Ratio and Association with Progression to End-Stage Renal Disease and Mortality. JAMA 2011, 305, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Gaspari, F.; Ruggenenti, P.; Porrini, E.; Motterlini, N.; Cannata, A.; Carrara, F.; Jiménez Sosa, A.; Cella, C.; Ferrari, S.; Stucchi, N.; et al. The GFR and GFR Decline Cannot Be Accurately Estimated in Type 2 Diabetics. Kidney Int. 2013, 84, 164–173. [Google Scholar] [CrossRef]
- Niwa, T.; Yamamoto, N.; Maeda, K.; Yamada, K.; Ohki, T.; Mori, M. Gas Chromatographic—Mass Spectrometric Analysis of Polyols in Urine and Serum of Uremic Patients: Identification of New Deoxyalditols and Inositol Isomers. J. Chromatogr. B Biomed. Sci. Appl. 1983, 277, 25–39. [Google Scholar] [CrossRef]
- Inamoto, Y.; Hiraga, Y.; Hanai, T.; Kinosita, T. The Development of a Sensitive Myo-Inositol Analyser Using a Liquid Chromatograph with a Post-Label Fluorescence Detector. Biomed. Chromatogr. 1995, 9, 146–149. [Google Scholar] [CrossRef]
- Michaelis, T.; Videen, J.S.; Linsey, M.S.; Ross, B.D. Dialysis and Transplantation Affect Cerebral Abnormalities of End-Stage Renal Disease. J. Magn. Reson. Imaging 1996, 6, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-Y.; Yoon, Y.J.; Choi, H.-J.; Park, S.-H.; Kim, C.-D.; Kim, I.-S.; Kwon, T.-H.; Do, J.-Y.; Kim, S.-H.; Ryu, D.H.; et al. Dialysis Modality-Dependent Changes in Serum Metabolites: Accumulation of Inosine and Hypoxanthine in Patients on Haemodialysis. Nephrol. Dial. Transplant. 2011, 26, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Ouyang, X.; Wang, L.; Peng, W.; Wen, J.; Dai, Y. A Pilot Metabolic Profiling Study in Serum of Patients with Chronic Kidney Disease Based on (1) H-NMR-Spectroscopy. Clin. Transl. Sci. 2012, 5, 379–385. [Google Scholar] [CrossRef]
- Sui, W.; Li, L.; Che, W.; Zuo, G.; Chen, J.; Li, W.; Dai, Y. A Proton Nuclear Magnetic Resonance-Based Metabonomics Study of Metabolic Profiling in Immunoglobulin a Nephropathy. Clinics 2012, 67, 363–373. [Google Scholar] [CrossRef]
- Al-Ani, B.; Fitzpatrick, M.; Al-Nuaimi, H.; Coughlan, A.M.; Hickey, F.B.; Pusey, C.D.; Savage, C.; Benton, C.M.; O’Brien, E.C.; O’Toole, D.; et al. Changes in Urinary Metabolomic Profile during Relapsing Renal Vasculitis. Sci. Rep. 2016, 6, 38074. [Google Scholar] [CrossRef] [PubMed]
- Duranton, F.; Lundin, U.; Gayrard, N.; Mischak, H.; Aparicio, M.; Mourad, G.; Daurès, J.-P.; Weinberger, K.M.; Argilés, A. Plasma and Urinary Amino Acid Metabolomic Profiling in Patients with Different Levels of Kidney Function. Clin. J. Am. Soc. Nephrol. 2014, 9, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Liu, X.; Wang, W.; Ren, H.; Xie, J.; Shen, P.; Lin, D.; Chen, N. Distinct Metabolic Profile of Primary Focal Segmental Glomerulosclerosis Revealed by NMR-Based Metabolomics. PLoS ONE 2013, 8, e78531. [Google Scholar] [CrossRef] [PubMed]
- Kraut, J.A.; Kurtz, I. Metabolic Acidosis of CKD: Diagnosis, Clinical Characteristics, and Treatment. Am. J. Kidney Dis. 2005, 45, 978–993. [Google Scholar] [CrossRef]
- Kumar, M.A.; Bitla, A.R.R.; Raju, K.V.N.; Manohar, S.M.; Kumar, V.S.; Narasimha, S.R.P.V.L. Branched Chain Amino Acid Profile in Early Chronic Kidney Disease. Saudi J. Kidney Dis. Transpl. 2012, 23, 1202–1207. [Google Scholar] [CrossRef]
- Mao, Y.-Y.; Bai, J.-Q.; Chen, J.-H.; Shou, Z.-F.; He, Q.; Wu, J.-Y.; Chen, Y.; Cheng, Y.-Y. A Pilot Study of GC/MS-Based Serum Metabolic Profiling of Acute Rejection in Renal Transplantation. Transpl. Immunol. 2008, 19, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Schnaper, H.W.; Furth, S.L.; Yao, L.P. Defining New Surrogate Markers for CKD Progression. Pediatr. Nephrol. 2015, 30, 193–198. [Google Scholar] [CrossRef]
| Development Set | Internal Validation Set | External Validation Set | p-Value | |
|---|---|---|---|---|
| N | 816 | 439 | 600 | |
| Age in years, mean ± SD (range) | 55 ± 14 (18–86) | 58 ± 15 (18–88) | 56 ± 14 (19–88) | 0.0269 1 |
| Height in cm, mean ± SD (range) | 170 ± 10 (142–199) | 170 ± 10 (143–199) 6 | 170 ± 10 (121–196) | 0.4929 1 |
| Weight in kg, mean ± SD (range) | 84 ± 21 (23–195) | 80 ± 19 (38–164) | 86 ± 21 (31–160) | <0.0001 1 |
| BMI in kg/m2, mean ± SD (range) | 29 ± 6 (7–68) | 28 ± 6 (15–52) 6 | 30 ± 6 (16–58) | 0.0005 1 |
| Male, N (%) | 431 (52.8%) | 143 (55.4%) | 334 (55.7%) | 0.5479 2 |
| Black, N (%) | 17 (2.1%) | 5 (1.1%) | 17 (2.8%) | 0.1575 2 |
| mGFR | ||||
| mean ± SD (range) 5 | 67 ± 28 (3–183) | 67 ± 28 (5–166) | 68 ± 27 (3–166) | 0.7280 1 |
| Iothalamate, N (%) | 816 (100.0%) | 269 (61.3%) | 600 (100.0%) | - 4 |
| Iohexol, N (%) | 0 (0.0%) | 92 (21.0%) | 0 (0.0%) | |
| Inulin, N (%) | 0 (0.0%) | 77 (17.5%) | 0 (0.0%) | |
| 51Cr-EDTA, N (%) | 0 (0.0%) | 1 (0.2%) | 0 (0.0%) | |
| CKD stage, N (%) | ||||
| G1 | 170 (20.8%) | 92 (21.0%) | 135 (22.5%) | 0.7989 3 |
| G2 | 291 (35.7%) | 156 (35.5%) | 214 (35.7%) | |
| G3 | 304 (37.3%) | 158 (36.0%) | 211 (35.1%) | |
| G4 | 46 (5.6%) | 26 (5.9%) | 36 (6.0%) | |
| G5 | 5 (0.6%) | 7 (1.6%) | 4 (0.7%) | |
| eGFR | ||||
| GFRNMR, mean ± SD (range) 5 | 66 ± 25 (7–151) | 66 ± 26 (7–175) | 67 ± 25 (9–158) | 0.5685 1 |
| CKD-EPI2009, mean ± SD (range) 5 | 63 ± 26 (5–165) | 68 ± 28 (4–146) | 64 ± 24 (8–142) | 0.0027 1 |
| CKD-EPI2012, mean ± SD (range) 5 | 60 ± 26 (6–144) | 63 ± 27 (6–145) | 62 ± 25 (8–150) | 0.1261 1 |
| CKD-EPIcys, mean ± SD (range) 5 | 59 ± 28 (7–136) | 60 ± 28 (7–148) | 61 ± 28 (9–144) | 0.3902 1 |
| EKFC, mean ± SD (range) 5 | 61 ± 23 (6–130) | 65 ± 25 (5–136) | 62 ± 22 (8–120) | 0.0074 1 |
| Sex | Serum Cystatin C [mg/L] | GFRNMR Equation 1 |
|---|---|---|
| Female | <1.02 | 238 × cystatin C−0.4114 × creatinine−0.3798 × valine0.1628 × 0.9979myo_inositol × 0.9963age |
| ≥1.02 | 239 × cystatin C−0.6443 × creatinine−0.3798 × valine0.1628 × 0.9979myo_inositol × 0.9963age | |
| Male | <1.22 | 266 × cystatin C−0.5867 × creatinine−0.3798 × valine0.1628 × 0.9979myo_inositol × 0.9963age |
| ≥1.22 | 269 × cystatin C−0.6419 × creatinine−0.3798 × valine0.1628 × 0.9979myo_inositol × 0.9963age |
| Variable | Estimated GFR Range | |||
|---|---|---|---|---|
| Overall | <60 | 60–89 | ≥90 | |
| n = 600 | mL/min/1.73 m2 of Body-Surface Area | |||
| Bias—median difference (95% CI) [mL/min/1.73 m2] | ||||
| CKD-EPI2009 | −4.0 (−4.8; −2.9) **** | −4.6 (−5.8; −3.3) | −4.0 (−6.2; −1.7) | −2.3 (−5.3; 1.1) |
| CKD-EPI2012 | −6.0 (−7.0; −5.0) **** | −7.0 (−8.0; −5.0) | −7.0 (−9.0; −4.0) | −2.0 (−6.0; 2.0) |
| CKD-EPICys | −7.0 (−8.1; −5.8) **** | −10.1 (−11.5; −8.2) | −5.8 (−7.9; −2.7) | 4.5 (−1; 7.5) |
| EKFC | −6.5 (−8.0; −5.0) **** | −6.0 (−8.0; −5.0) | −8.0 (−10.0; −5.0) | −4.0 (−10.0; 2.0) |
| GFRNMR | 0.0 (−1.0; 1.0) | −1.0 (−2.0; 1.0) | 0.0 (−3.0; 1.0) | 3.0 (0.0; 5.0) |
| Precision—IQR of the difference (95% CI) [mL/min/1.73 m2] | ||||
| CKD-EPI2009 | 16.4 (15.1; 17.7) * | 14.4 (12.5; 15.9) | 18.5 (15.5; 22.3) | 25.6 (20.4; 34.6) |
| CKD-EPI2012 | 14.0 (12.0; 16.0) | 11.0 (10.0; 13.0) | 16.0 (13.0; 19.0) | 21.5 (14.5; 27.5) |
| CKD-EPICys | 18.1 (16.4; 19.9) * | 14.4 (12.4; 16.1) | 20.4 (16.3; 22.9) | 23.3 (15.5; 27.3) |
| EKFC | 17.0 (15.0; 19.0) * | 15.0 (12.0; 16.0) | 20.0 (17.0; 24.0) | 32.0 (22.0; 42.0) |
| GFRNMR | 13.0 (10.0; 14.0) | 11.0 (10.0; 13.0) | 15.0 (13.0; 19.0) | 20.0 (15.0; 26.0) |
| Error—mean absolute error (95% CI) [mL/min/1.73 m2] | ||||
| CKD-EPI2009 | 11.9 (11.0; 12.7) **** | 9.4 (8.5; 10.4) | 12.8 (11.3; 14.4) | 16.6 (14.2; 19.3) |
| CKD-EPI2012 | 11.1 (10.3; 11.9) **** | 9.6 (8.8; 10.6) | 12.0 (10.6; 13.5) | 14.0 (11.7; 16.5) |
| CKD-EPICys | 13.3 (12.3; 14.2) **** | 12.9 (11.7; 14.1) | 13.2 (11.4; 14.9) | 14.4 (12.1; 16.9) |
| EKFC | 12.8 (11.9; 13.7) **** | 10.3 (9.3; 11.2) | 14.3 (12.6; 16.1) | 18.5 (15.5; 21.6) |
| GFRNMR | 10.0 (9.2; 10.8) | 7.6 (6.8; 8.5) | 10.6 (9.4; 11.8) | 14.1 (11.9; 16.4) |
| Accuracy—1-P15 (95% CI) [%] | ||||
| CKD-EPI2009 | 49.5 (45.0; 53.7) **** | 54.1 (48.6; 60) | 44.4 (37.9; 51.0) | 46.4 (37.5; 55.4) |
| CKD-EPI2012 | 47.3 (43.2; 51.5) ** | 57.7 (52.3; 63.2) | 37.7 (30.9; 45.0) | 33.3 (24.2; 43.4) |
| CKD-EPICys | 56.0 (51.8; 60.0) **** | 68.3 (63.1; 73.5) | 43.4 (35.2; 51.6) | 38.1 (29.2; 46.9) |
| EKFC | 53.0 (49.0; 57.0) **** | 57.7 (52.1; 63.2) | 46.8 (40.3; 53.7) | 51.9 (40.3; 62.3) |
| GFRNMR | 38.8 (34.3; 42.5) | 43.5 (37.4; 49.6) | 35.4 (29.5; 41.4) | 35.9 (27.4; 44.4) |
| Accuracy—1-P20 (95% CI) [%] | ||||
| CKD-EPI2009 | 37.2 (33; 41.3) *** | 42.1 (36.6; 47.9) | 30.3 (24.2; 36.4) | 36.6 (28.6; 45.5) |
| CKD-EPI2012 | 33.3 (29.3; 37.2) * | 41.9 (36.1; 47.4) | 24.6 (18.8; 30.9) | 23.2 (15.2; 32.3) |
| CKD-EPICys | 44.0 (39.7; 48.0) **** | 57.0 (51.5; 62.5) | 32.1 (24.5; 39.6) | 23.0 (15.9; 31.0) |
| EKFC | 40.5 (36.3; 44.7) **** | 45.3 (39.7; 51.1) | 33.3 (26.9; 40.3) | 41.6 (31.2; 51.9) |
| GFRNMR | 28.5 (24.7; 32.2) | 32.1 (26.4; 37.8) | 26.2 (20.7; 32.1) | 25.6 (17.9; 34.2) |
| Accuracy—1-P30 (95% CI) [%] | ||||
| CKD-EPI2009 | 15.5 (12.5; 18.7) | 17.6 (13.4; 22.1) | 13.1 (8.6; 18.2) | 14.3 (8.0; 21.4) |
| CKD-EPI2012 | 13.2 (10.2; 16.0) | 17.7 (13.5; 22.3) | 7.9 (4.2; 12.0) | 9.1 (4.0; 15.2) |
| CKD-EPICys | 22.8 (19.3; 26.2) **** | 32.6 (27.4; 38.1) | 11.9 (6.9; 17.0) | 9.7 (4.4; 15.9) |
| EKFC | 16.3 (13.2; 19.5) | 18.9 (14.7; 23.4) | 13.9 (9.3; 18.5) | 13.0 (5.2; 20.8) |
| GFRNMR | 12.8 (9.8; 15.5) | 17.5 (13.0; 22.4) | 9.3 (5.5; 13.1) | 10.3 (5.1; 16.2) |
| Estimated GFR Range (mL/min/1.73 m2) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Overall | <15 | 15–29 | 30–44 | 45–59 | 60–89 | ≥90 | ||
| CKD stage | - | G5 | G4 | G3b | G3a | G2 | G1 | |
| Observed mGFR range (mL/min/1.73 m2) | 3–166 | 3–14 | 16–29 | 30–44 | 45–59 | 60–89 | 90–166 | |
| Number of samples | 600 | 4 | 36 | 79 | 132 | 214 | 135 | |
| CKD-EPI2009 | Total reclassified, N (%) | 218 (36.4) | 0 (0.0) | 10 (27.8) | 19 (24.1) | 64 (48.5) | 76 (35.5) | 49 (36.3) |
| Correctly reclassified, N (%) 1 | 133 (22.2) | 0 (0.0) | 6 (16.7) | 8 (10.1) | 38 (28.8) | 54 (25.2) | 27 (20.0) | |
| Incorrectly reclassified, N (%) 2 | 85 (14.2) | 0 (0.0) | 4 (11.1) | 11 (13.9) | 26 (19.7) | 22 (10.3) | 22 (16.3) | |
| NRI (%) | 8.0 | 0.0 | 5.6 | −3.8 | 9.1 | 14.9 | 3.7 | |
| CKD-EPI2012 | Total reclassified, N (%) | 168 (28.0) | 1 (25.0) | 8 (22.2) | 32 (40.5) | 59 (44.7) | 49 (22.9) | 19 (14.1) |
| Correctly reclassified, N (%) 1 | 107 (17.8) | 0 (0) | 1 (2.8) | 16 (20.3) | 36 (27.3) | 40 (18.7) | 14 (10.4) | |
| Incorrectly reclassified, N (%) 2 | 61 (10.2) | 1 (25.0) | 7 (19.4) | 16 (20.3) | 23 (17.4) | 9 (4.2) | 5 (3.7) | |
| NRI (%) | 7.6 | −25.0 | −16.6 | 0.0 | 9.9 | 14.5 | 6.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stämmler, F.; Grassi, M.; Meeusen, J.W.; Lieske, J.C.; Dasari, S.; Dubourg, L.; Lemoine, S.; Ehrich, J.; Schiffer, E. Estimating Glomerular Filtration Rate from Serum Myo-Inositol, Valine, Creatinine and Cystatin C. Diagnostics 2021, 11, 2291. https://doi.org/10.3390/diagnostics11122291
Stämmler F, Grassi M, Meeusen JW, Lieske JC, Dasari S, Dubourg L, Lemoine S, Ehrich J, Schiffer E. Estimating Glomerular Filtration Rate from Serum Myo-Inositol, Valine, Creatinine and Cystatin C. Diagnostics. 2021; 11(12):2291. https://doi.org/10.3390/diagnostics11122291
Chicago/Turabian StyleStämmler, Frank, Marcello Grassi, Jeffrey W. Meeusen, John C. Lieske, Surendra Dasari, Laurence Dubourg, Sandrine Lemoine, Jochen Ehrich, and Eric Schiffer. 2021. "Estimating Glomerular Filtration Rate from Serum Myo-Inositol, Valine, Creatinine and Cystatin C" Diagnostics 11, no. 12: 2291. https://doi.org/10.3390/diagnostics11122291
APA StyleStämmler, F., Grassi, M., Meeusen, J. W., Lieske, J. C., Dasari, S., Dubourg, L., Lemoine, S., Ehrich, J., & Schiffer, E. (2021). Estimating Glomerular Filtration Rate from Serum Myo-Inositol, Valine, Creatinine and Cystatin C. Diagnostics, 11(12), 2291. https://doi.org/10.3390/diagnostics11122291





