Clinical Evaluation of a New Antigen-Based COVID-19 Rapid Diagnostic Test from Symptomatic Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Sample Collection
2.3. Onsite® Rapid Test
2.4. RT-PCR for COVID-19 Diagnosis
2.5. Sanger Sequencing and Clade Identification
2.6. Ethical Considerations
2.7. Data Analysis
3. Results
3.1. Enrollment
3.2. COVID-19 Patients Confirmed by RT-PCR
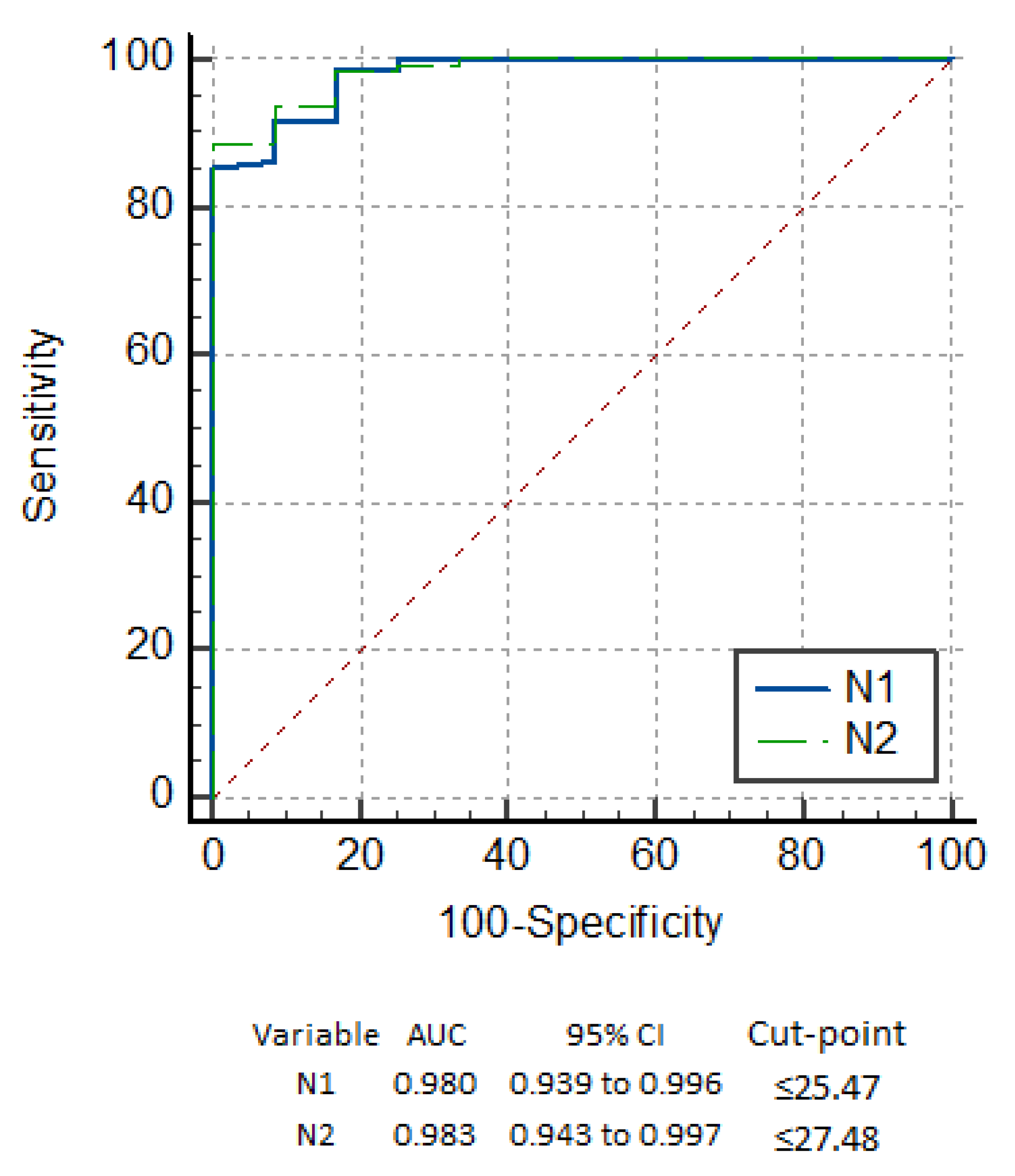
3.3. Performance of the Onsite COVID Ag RDT
3.4. Detection Based on Symptoms
3.5. Detection Based on Variants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pascarella, G.; Strumia, A.; Piliego, C.; Bruno, F.; Del Buono, R.; Costa, F.; Scarlata, S.; Agrò, F.E. COVID-19 diagnosis and management: A comprehensive review. J. Intern. Med. 2020, 288, 192–206. [Google Scholar] [CrossRef]
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 5 August 2021).
- West, C.P.; Montori, V.M.; Sampathkumar, P. COVID-19 Testing: The threat of false-negative results. Mayo Clin. Proc. 2020, 95, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Li, R.; Han, Y.; Zhang, R.; Li, J. COVID-19: Insight into the asymptomatic SARS-COV-2 infection and transmission. Int. J. Biol. Sci. 2020, 16, 2803–2811. [Google Scholar] [CrossRef] [PubMed]
- Nikolai, L.A.; Meyer, C.G.; Kremsner, P.G.; Velavan, T.P. Asymptomatic SARS Coronavirus 2 infection: Invisible yet invincible. Int. J. Infect. Dis. 2020, 100, 112–116. [Google Scholar] [CrossRef]
- Wouters, O.J.; Shadlen, K.C.; Salcher-Konrad, M.; Pollard, A.J.; Larson, H.J.; Teerawattananon, Y.; Jit, M. Challenges in ensuring global access to COVID-19 vaccines: Production, affordability, allocation, and deployment. Lancet 2021, 397, 1023–1034. [Google Scholar] [CrossRef]
- Trogen, B.; Pirofski, L.-A. Understanding vaccine hesitancy in COVID-19. Medicine 2021, 2, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Klaser, K.; May, A.; Polidori, L.; Capdevila, J.; Louca, P.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Merino, J.; et al. Vaccine side-effects and SARS-CoV-2 infection after vaccination in users of the COVID Symptom Study app in the UK: A prospective observational study. Lancet. Infect. Dis. 2021, 21, 939–949. [Google Scholar] [CrossRef]
- Popp, M.; Stegemann, M.; Metzendorf, M.I.; Gould, S.; Kranke, P.; Meybohm, P.; Skoetz, N.; Weibel, S. Ivermectin for preventing and treating COVID-19. Cochrane Database Syst. Rev. 2021, 2021, CD015017. [Google Scholar] [CrossRef]
- Rodriguez-Guerra, M.; Jadhav, P.; Vittorio, T.J. Current treatment in COVID-19 disease: A rapid review. Drugs Context 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ayeh, S.K.; Chidambaram, V.; Karakousis, P.C. Modes of transmission of SARS-CoV-2 and evidence for preventive behavioral interventions. BMC Infect. Dis. 2021, 21, 496. [Google Scholar] [CrossRef] [PubMed]
- Lokuge, K.; Banks, E.; Davis, S.; Roberts, L.; Street, T.; O’Donovan, D.; Caleo, G.; Glass, K. Exit strategies: Optimising feasible surveillance for detection, elimination, and ongoing prevention of COVID-19 community transmission. BMC Med. 2021, 19, 50. [Google Scholar] [CrossRef]
- Khan, A.A.; Alahdal, H.M.; Alotaibi, R.M.; Sonbol, H.S.; Almaghrabi, R.H.; Alsofayan, Y.M.; Althunayyan, S.M.; Alsaif, F.A.; Almudarra, S.S.; Alabdulkareem, K.I.; et al. Controlling COVID-19 Pandemic: A Mass Screening Experience in Saudi Arabia. Front. Public Health 2021, 8, 606385. [Google Scholar] [CrossRef] [PubMed]
- Alkharsah, K.R. Laboratory tests for the detection of SARS-CoV-2 infection: Basic principles and examples. Ger. Med. Sci. 2021, 19, Doc06. [Google Scholar] [CrossRef] [PubMed]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 diagnosis-A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John Leon Singh, H.; Couch, D.; Yap, K. Mobile Health Apps that help with COVID-19 management: Scoping review. JMIR Nurs. 2020, 3, e20596. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Mishra, A.S. COVID-19 infection: Disease detection and mobile technology. PeerJ 2020, 8, e10345. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Augustine, S.; Narayan, T.; O‘Riordan, A.; Das, A.; Kumar, D.; Luong, J.H.T.; Malhotra, B.D. Point-of-Care PCR Assays for COVID-19 Detection. Biosensors 2021, 11, 141. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.N.; de Oliveira Coelho, B.; Góes, L.G.B.; Minoprio, P.; Durigon, E.L.; Morello, L.G.; Marchini, F.K.; Riediger, I.N.; do Carmo Debur, M.; Nakaya, H.I.; et al. Colorimetric RT-LAMP SARS-CoV-2 diagnostic sensitivity relies on color interpretation and viral load. Sci. Rep. 2021, 11, 9026. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi, S.; Fontana, S.; Mambrin, F.; Nguyen, H.Q.; Righi, E.; Tacconelli, E.; Mansueto, G. Pitfalls of Computed Tomography in the Coronavirus 2019 (COVID-19) Era: A New Perspective on Ground-Glass Opacities. Cureus 2020, 12, e8151. [Google Scholar] [CrossRef] [PubMed]
- Kudo, E.; Israelow, B.; Vogels, C.B.F.; Lu, P.; Wyllie, A.L.; Tokuyama, M.; Venkataraman, A.; Brackney, D.E.; Ott, I.M.; Petrone, M.E.; et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biol. 2020, 18, e3000867. [Google Scholar] [CrossRef] [PubMed]
- Quick, J. nCoV-2019 Sequencing Protocol v3 (LoCost). 2020. Available online: https://protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye (accessed on 19 September 2021).
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef] [PubMed]
- Saeed, U.; Uppal, S.R.; Piracha, Z.Z.; Rasheed, A.; Aftab, Z.; Zaheer, H.; Uppal, R. Evaluation of SARS-CoV-2 antigen-based rapid diagnostic kits in Pakistan: Formulation of COVID-19 national testing strategy. Virol. J. 2021, 18, 34. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef]
- Mak, G.C.K.; Lau, S.S.Y.; Wong, K.K.Y.; Chow, N.L.S.; Lau, C.S.; Lam, E.T.K.; Chan, R.C.W.; Tsang, D.N.C. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J. Clin. Virol. 2021, 134, 104712. [Google Scholar] [CrossRef]
- Cerutti, F.; Burdino, E.; Milia, M.G.; Allice, T.; Gregori, G.; Bruzzone, B.; Ghisetti, V. Urgent need of rapid tests for SARS CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 2020, 132, 104654. [Google Scholar] [CrossRef] [PubMed]
- Edson, D.C.; Casey, D.L.; Harmer, S.E.; Downes, F.P. Identification of SARS-CoV-2 in a Proficiency Testing Program. Am. J. Clin. Pathol. 2020, 154, 475–478. [Google Scholar] [CrossRef]
- Freire-Paspuel, B.; Garcia-Bereguiain, M.A. Analytical and Clinical Evaluation of “AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Bioneer, South Korea)” and “Allplex 2019-nCoV Assay (Seegene, South Korea)” for SARS-CoV-2 RT-PCR Diagnosis: Korean CDC EUA as a Quality Control Proxy for Developing Countries. Front. Cell. Infect. Microbiol. 2021, 11, 630552. [Google Scholar] [CrossRef]
- Berger, A.; Nsoga, M.T.N.; Perez-Rodriguez, F.J.; Aad, Y.A.; Sattonnet-Roche, P.; Gayet-Ageron, A.; Jaksic, C.; Torriani, G.; Boehm, E.; Kronig, I.; et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS ONE 2021, 16, e0248921. [Google Scholar] [CrossRef] [PubMed]
- Albert, E.; Torres, I.; Bueno, F.; Huntley, D.; Molla, E.; Fernández-Fuentes, M.; Martínez, M.; Poujois, S.; Forqué, L.; Valdivia, A.; et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021, 27, 472. [Google Scholar] [CrossRef] [PubMed]
- Torres, I.; Poujois, S.; Albert, E.; Colomina, J.; Navarro, D. Evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin. Microbiol. Infect. 2021, 27, 636. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.-H.; Park, K.; Lim, Y.; Jeong, Y.S.; Sung, H.; Kim, M.-N. Evaluation of Four Commercial Kits for SARS-CoV-2 Real-Time Reverse-Transcription Polymerase Chain Reaction Approved by Emergency-Use-Authorization in Korea. Front. Med. 2020, 7, 521. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Emergency Use Assessment Coronavirus Disease (COVID-19) IVDs, PUBLIC REPORT. 2020. Available online: https://www.who.int/diagnostics_laboratory/eual/201019_final_pqpr_eul_0563_117_00_standard_q_covid19_ag_test.pdf?ua=1 (accessed on 3 October 2021).
- Kweon, O.J.; Lim, Y.K.; Kim, H.R.; Choi, Y.; Kim, M.-C.; Choi, S.-H.; Chung, J.-W.; Lee, M.-K. Evaluation of rapid SARS-CoV-2 antigen tests, AFIAS COVID-19 Ag and ichroma COVID-19 Ag, with serial nasopharyngeal specimens from COVID-19 patients. PLoS ONE 2021, 16, e0249972. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sazed, S.A.; Kibria, M.G.; Hossain, M.S.; Zamil, M.F.; Adhikary, P.C.; Hossain, M.E.; Ahmed, D.; Haque, R.; Alam, M.S. Clinical Evaluation of a New Antigen-Based COVID-19 Rapid Diagnostic Test from Symptomatic Patients. Diagnostics 2021, 11, 2300. https://doi.org/10.3390/diagnostics11122300
Sazed SA, Kibria MG, Hossain MS, Zamil MF, Adhikary PC, Hossain ME, Ahmed D, Haque R, Alam MS. Clinical Evaluation of a New Antigen-Based COVID-19 Rapid Diagnostic Test from Symptomatic Patients. Diagnostics. 2021; 11(12):2300. https://doi.org/10.3390/diagnostics11122300
Chicago/Turabian StyleSazed, Saiful Arefeen, Mohammad Golam Kibria, Mohammad Sharif Hossain, Md Fahad Zamil, Pranob Chandra Adhikary, Mohammad Enayet Hossain, Dilruba Ahmed, Rashidul Haque, and Mohammad Shafiul Alam. 2021. "Clinical Evaluation of a New Antigen-Based COVID-19 Rapid Diagnostic Test from Symptomatic Patients" Diagnostics 11, no. 12: 2300. https://doi.org/10.3390/diagnostics11122300
APA StyleSazed, S. A., Kibria, M. G., Hossain, M. S., Zamil, M. F., Adhikary, P. C., Hossain, M. E., Ahmed, D., Haque, R., & Alam, M. S. (2021). Clinical Evaluation of a New Antigen-Based COVID-19 Rapid Diagnostic Test from Symptomatic Patients. Diagnostics, 11(12), 2300. https://doi.org/10.3390/diagnostics11122300






