Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort and Data Collection
2.2. Analysis of Altered Signaling Pathways
2.3. Comparison of Datasets from NCKUH and MSKCC Cohort
2.4. Statistical Analysis
3. Results
3.1. Characteristics of NCKUH Cohort
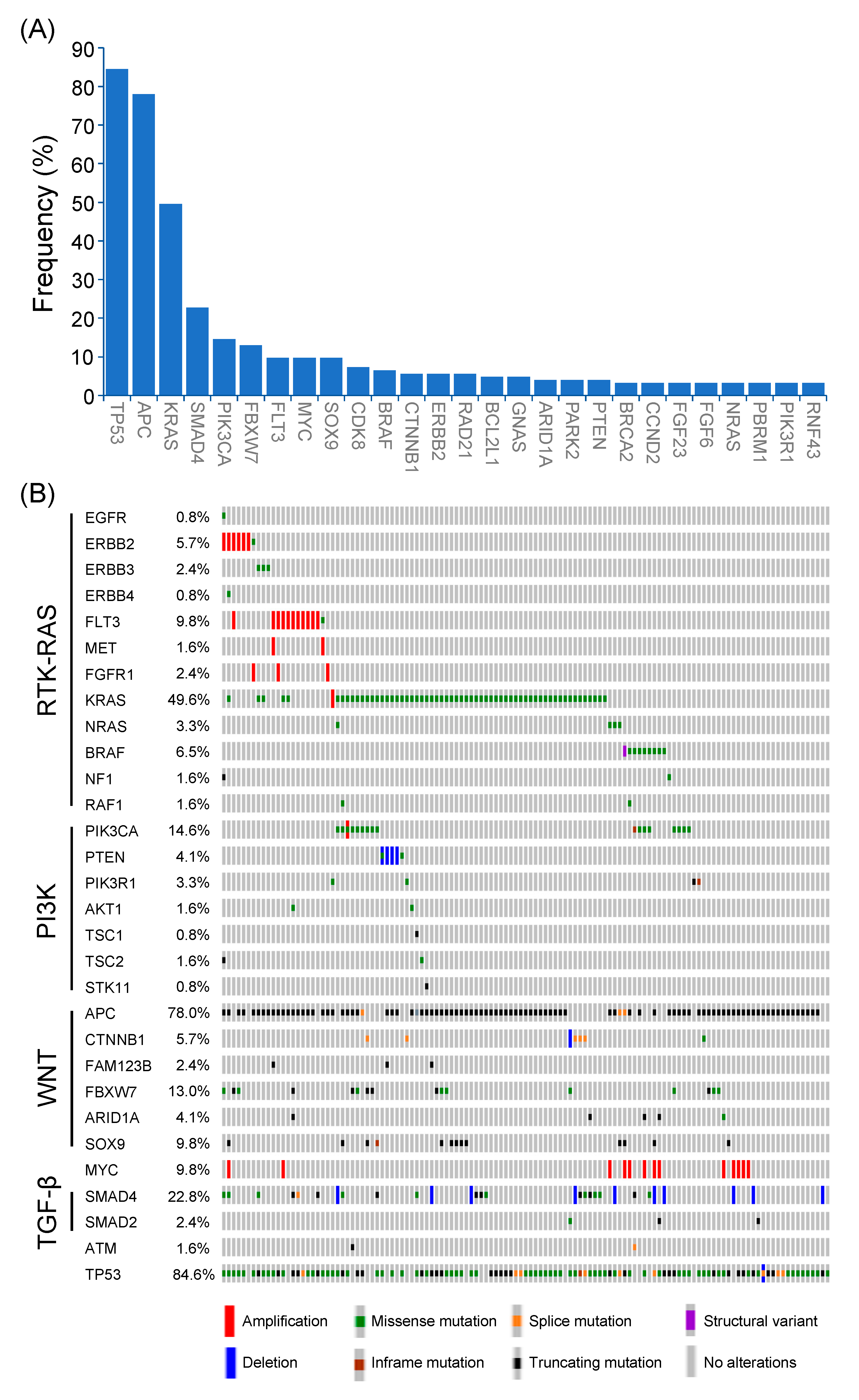
3.2. Genomic Alterations in Non-MSI-H CRC
3.3. Alterations of Signaling Pathways
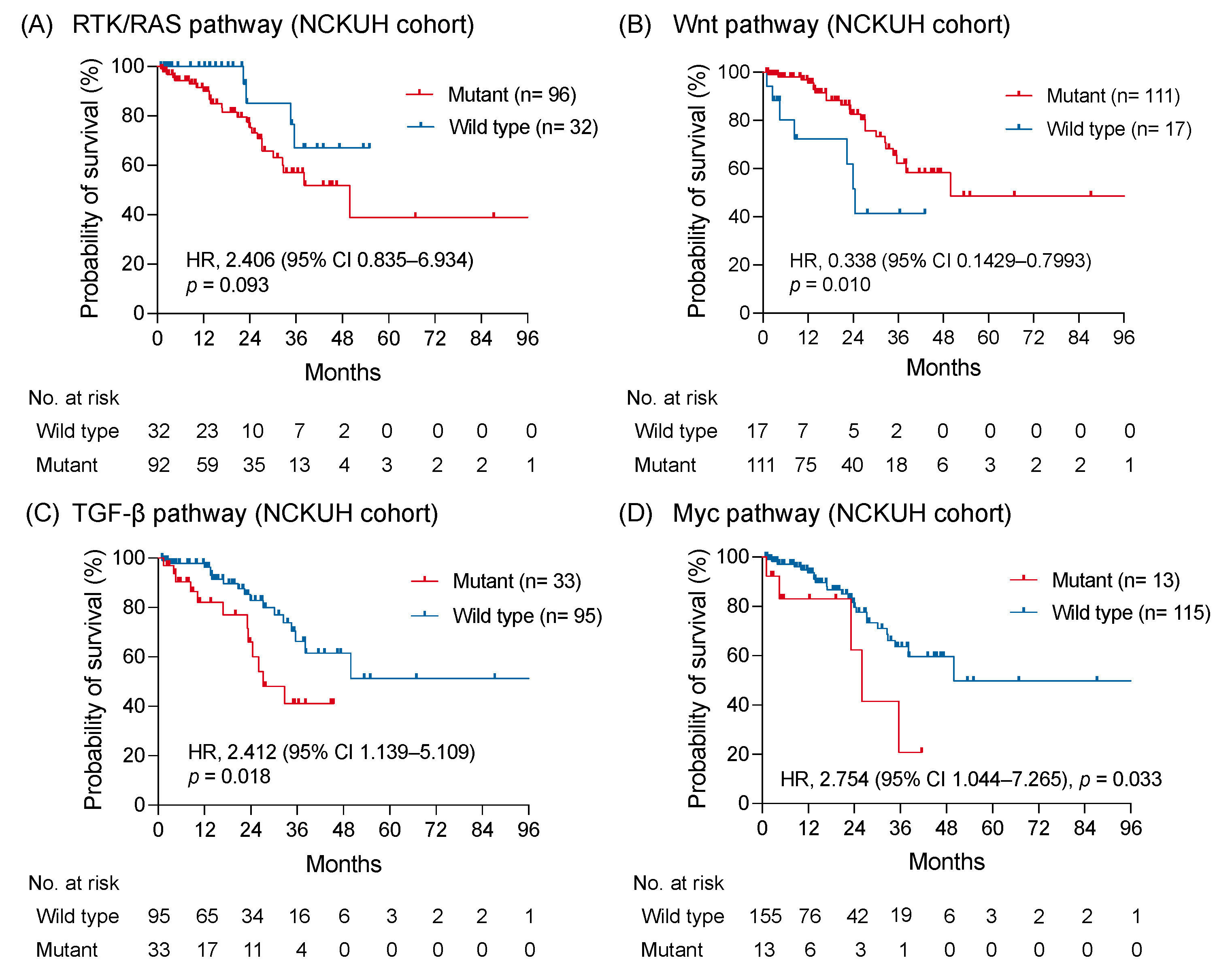
3.4. Clinical Outcome of Signaling Pathway with Alteration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- Bockelman, C.; Engelmann, B.E.; Kaprio, T.; Hansen, T.F.; Glimelius, B. Risk of recurrence in patients with colon cancer stage II and III: A systematic review and meta-analysis of recent literature. Acta Oncol. 2015, 54, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Sugarbaker, P.H. Colorectal cancer: Prevention and management of metastatic disease. BioMed Res. Int. 2014, 2014, 782890. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Isoniemi, H.; Uutela, A.; Nordin, A.; Lantto, E.; Kellokumpu, I.; Ovissi, A.; Kosunen, J.; Kallio, R.; Soveri, L.M.; Salminen, T.; et al. Centralized repeated resectability assessment of patients with colorectal liver metastases during first-line treatment: Prospective study. Br. J. Surg. 2021, 108, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Saraf, S.; Verma, A.; Panda, P.K.; Jain, S.K. Novel targeting approaches and signaling pathways of colorectal cancer: An insight. World J. Gastroenterol. 2018, 24, 4428–4435. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Singh, J.K.; Wunnava, A.; Al-Obeed, O.; Abdulla, M.; Srivastava, S.K. Emerging trends in colorectal cancer: Dysregulated signaling pathways (Review). Int. J. Mol. Med. 2021, 47, 14. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Tabernero, J. Spectrum of Gene Mutations in Colorectal Cancer: Implications for Treatment. Cancer J. 2016, 22, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Salazar, R.; Tabernero, J. Molecular Subtypes and the Evolution of Treatment Decisions in Metastatic Colorectal Cancer. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 231–238. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Duong, H.Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy. Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018, 33, 125–136.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FoundationOne CDx Home Page. Available online: https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx (accessed on 10 August 2021).
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Bahceci, I.; Dogrusoz, U.; La, K.C.; Babur, O.; Gao, J.; Schultz, N. PathwayMapper: A collaborative visual web editor for cancer pathways and genomic data. Bioinformatics 2017, 33, 2238–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef]
- Zhou, Z.; Xie, X.; Wang, X.; Zhang, X.; Li, W.; Sun, T.; Cai, Y.; Wu, J.; Dang, C.; Zhang, H. Correlations Between Tumor Mutation Burden and Immunocyte Infiltration and Their Prognostic Value in Colon Cancer. Front. Genet. 2021, 12, 623424. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Jung, B.; Staudacher, J.J.; Beauchamp, D. Transforming Growth Factor beta Superfamily Signaling in Development of Colorectal Cancer. Gastroenterology 2017, 152, 36–52. [Google Scholar] [CrossRef] [Green Version]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-beta Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.L.; Chen, Z.Q.; Zhu, S.L.; Liu, T.W.; Wen, Y.; Su, Y.S.; Xi, X.J.; Hu, Y.; Lian, L.; Liu, F.B. Prognostic value of transforming growth factor-beta in patients with colorectal cancer who undergo surgery: A meta-analysis. BMC Cancer 2017, 17, 240. [Google Scholar] [CrossRef]
- Misale, S.; Di Nicolantonio, F.; Sartore-Bianchi, A.; Siena, S.; Bardelli, A. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 2014, 4, 1269–1280. [Google Scholar] [CrossRef] [Green Version]
- Sforza, V.; Martinelli, E.; Ciardiello, F.; Gambardella, V.; Napolitano, S.; Martini, G.; Della Corte, C.; Cardone, C.; Ferrara, M.L.; Reginelli, A.; et al. Mechanisms of resistance to anti-epidermal growth factor receptor inhibitors in metastatic colorectal cancer. World J. Gastroenterol. 2016, 22, 6345–6361. [Google Scholar] [CrossRef]
- Parseghian, C.M.; Napolitano, S.; Loree, J.M.; Kopetz, S. Mechanisms of Innate and Acquired Resistance to Anti-EGFR Therapy: A Review of Current Knowledge with a Focus on Rechallenge Therapies. Clin. Cancer Res. 2019, 25, 6899–6908. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef]
- Bedi, A.; Chang, X.; Noonan, K.; Pham, V.; Bedi, R.; Fertig, E.J.; Considine, M.; Califano, J.A.; Borrello, I.; Chung, C.H.; et al. Inhibition of TGF-beta enhances the in vivo antitumor efficacy of EGF receptor-targeted therapy. Mol. Cancer Ther. 2012, 11, 2429–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Zhang, L.; Zhou, J.; Zheng, J. Silencing Smad4 attenuates sensitivity of colorectal cancer cells to cetuximab by promoting epithelialmesenchymal transition. Mol. Med. Rep. 2019, 20, 3735–3745. [Google Scholar] [CrossRef] [Green Version]
- Huynh, L.K.; Hipolito, C.J.; Ten Dijke, P. A Perspective on the Development of TGF-beta Inhibitors for Cancer Treatment. Biomolecules 2019, 9, 743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciardiello, D.; Elez, E.; Tabernero, J.; Seoane, J. Clinical development of therapies targeting TGFbeta: Current knowledge and future perspectives. Ann. Oncol. 2020, 31, 1336–1349. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Chung, C.L.; Hu, T.H.; Chen, J.J.; Liu, P.F.; Chen, C.L. Recent progress in TGF-beta inhibitors for cancer therapy. Biomed. Pharmacother. 2021, 134, 111046. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Dang, C.V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a014217. [Google Scholar] [CrossRef] [PubMed]
- Bretones, G.; Delgado, M.D.; Leon, J. Myc and cell cycle control. Biochim. Biophys. Acta 2015, 1849, 506–516. [Google Scholar] [CrossRef]
- Toon, C.W.; Chou, A.; Clarkson, A.; DeSilva, K.; Houang, M.; Chan, J.C.; Sioson, L.L.; Jankova, L.; Gill, A.J. Immunohistochemistry for myc predicts survival in colorectal cancer. PLoS ONE 2014, 9, e87456. [Google Scholar] [CrossRef]
- Lee, K.S.; Kwak, Y.; Nam, K.H.; Kim, D.W.; Kang, S.B.; Choe, G.; Kim, W.H.; Lee, H.S. c-MYC Copy-Number Gain Is an Independent Prognostic Factor in Patients with Colorectal Cancer. PLoS ONE 2015, 10, e0139727. [Google Scholar] [CrossRef]
- Lee, K.S.; Kwak, Y.; Nam, K.H.; Kim, D.W.; Kang, S.B.; Choe, G.; Kim, W.H.; Lee, H.S. Favorable prognosis in colorectal cancer patients with co-expression of c-MYC and ss-catenin. BMC Cancer 2016, 16, 730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, W.L.; Weng, X.T.; Wang, J.L.; Lin, Y.K.; Liu, T.W.; Zhou, Q.Y.; Hu, Y.; Pan, Y.; Chen, X.L. Association Between c-Myc and Colorectal Cancer Prognosis: A Meta-Analysis. Front. Physiol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, T.; Matsuhashi, N.; Tomita, H.; Takahashi, T.; Iwata, Y.; Fukada, M.; Yasufuku, I.; Suetsugu, T.; Imai, T.; Mori, R.; et al. MYC Up-regulation Is a Useful Biomarker for Preoperative Neoadjuvant Chemotherapy Combined with Anti-EGFR in Liver Metastasis from Colorectal Cancer. In Vivo 2021, 35, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, B.; Carlsson, G.; Machover, D.; Petrelli, N.; Roth, A.; Schmoll, H.J.; Tveit, K.M.; Gibson, F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin. Colorectal Cancer 2015, 14, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McQuade, R.M.; Stojanovska, V.; Bornstein, J.C.; Nurgali, K. Colorectal Cancer Chemotherapy: The Evolution of Treatment and New Approaches. Curr. Med. Chem. 2017, 24, 1537–1557. [Google Scholar] [CrossRef]
- Prochownik, E.V. c-Myc: Linking transformation and genomic instability. Curr. Mol. Med. 2008, 8, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Pyndiah, S.; Tanida, S.; Ahmed, K.M.; Cassimere, E.K.; Choe, C.; Sakamuro, D. c-MYC suppresses BIN1 to release poly(ADP-ribose) polymerase 1: A mechanism by which cancer cells acquire cisplatin resistance. Sci. Signal. 2011, 4, ra19. [Google Scholar] [CrossRef]
- Kuzyk, A.; Mai, S. c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 2014, 4, a014373. [Google Scholar] [CrossRef]
- Reyes-Gonzalez, J.M.; Armaiz-Pena, G.N.; Mangala, L.S.; Valiyeva, F.; Ivan, C.; Pradeep, S.; Echevarria-Vargas, I.M.; Rivera-Reyes, A.; Sood, A.K.; Vivas-Mejia, P.E. Targeting c-MYC in Platinum-Resistant Ovarian Cancer. Mol. Cancer Ther. 2015, 14, 2260–2269. [Google Scholar] [CrossRef] [Green Version]
- Seligmann, J.F.; Fisher, D.; Smith, C.G.; Richman, S.D.; Elliott, F.; Brown, S.; Adams, R.; Maughan, T.; Quirke, P.; Cheadle, J.; et al. Investigating the poor outcomes of BRAF-mutant advanced colorectal cancer: Analysis from 2530 patients in randomised clinical trials. Ann. Oncol. 2017, 28, 562–568. [Google Scholar] [CrossRef]
- Sanz-Garcia, E.; Argiles, G.; Elez, E.; Tabernero, J. BRAF mutant colorectal cancer: Prognosis, treatment, and new perspectives. Ann. Oncol. 2017, 28, 2648–2657. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [Green Version]
- Molina-Cerrillo, J.; San Roman, M.; Pozas, J.; Alonso-Gordoa, T.; Pozas, M.; Conde, E.; Rosas, M.; Grande, E.; Garcia-Bermejo, M.L.; Carrato, A. BRAF Mutated Colorectal Cancer: New Treatment Approaches. Cancers 2020, 12, 1571. [Google Scholar] [CrossRef] [PubMed]
- Barras, D.; Missiaglia, E.; Wirapati, P.; Sieber, O.M.; Jorissen, R.N.; Love, C.; Molloy, P.L.; Jones, I.T.; McLaughlin, S.; Gibbs, P.; et al. BRAF V600E Mutant Colorectal Cancer Subtypes Based on Gene Expression. Clin. Cancer Res. 2017, 23, 104–115. [Google Scholar] [CrossRef] [Green Version]
- Strippoli, A.; Cocomazzi, A.; Basso, M.; Cenci, T.; Ricci, R.; Pierconti, F.; Cassano, A.; Fiorentino, V.; Barone, C.; Bria, E.; et al. c-MYC Expression Is a Possible Keystone in the Colorectal Cancer Resistance to EGFR Inhibitors. Cancers 2020, 12, 638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grbcic, P.; Fuckar Cupic, D.; Gamberi, T.; Kraljevic Pavelic, S.; Sedic, M. Proteomic Profiling of BRAFV600E Mutant Colon Cancer Cells Reveals the Involvement of Nucleophosmin/c-Myc Axis in Modulating the Response and Resistance to BRAF Inhibition by Vemurafenib. Int. J. Mol. Sci. 2021, 22, 6174. [Google Scholar] [CrossRef] [PubMed]



| Characteristic | All Subjects | MSI-H | Non-MSI-H |
|---|---|---|---|
| Total patients | 128 | 5 | 123 |
| Age—yr | |||
| Median | 61 | 47 | 61 |
| Range | 26–87 | 37–66 | 26–87 |
| Gender—no. (%) | |||
| Male | 76 (59.4) | 4 (80.0) | 72 (58.5) |
| Female | 52 (40.6) | 1 (20.0) | 51 (41.5) |
| Primary tumor location—no. (%) | |||
| Right-sided | 29 (22.7) | 4 (80.0) | 25 (20.3) |
| Left-sided | 99 (77.3) | 1 (20.0) | 98 (79.7) |
| Tumor sample for testing—no. (%) | |||
| Primary lesion | 71 (55.5) | 3 (60.0) | 68 (55.3) |
| Metastatic lesion | 57 (44.5) | 2 (40.0) | 55 (44.7) |
| Tumor histology—no. (%) | |||
| Adenocarcinoma | 122 (95.3) | 5 (100) | 117 (95.1) |
| Mucinous adenocarcinoma | 6 (4.7) | 0 | 6 (4.9) |
| Tumor grade—no. (%) | |||
| Well differentiated | 5 (3.9) | 0 | 5 (4.1) |
| Moderately differentiated | 102 (79.7) | 4 (80.0) | 98 (79.7) |
| Poorly differentiated | 12 (9.4) | 1 (20.0) | 11 (8.9) |
| NA | 9 (7.0) | 0 | 9 (7.3) |
| First-line regimen—no. (%) | |||
| FOLFIRI + Bevacizumab | 75 (58.6) | 3 (60.0) | 72 (58.5) |
| FOLFIRI + Cetuximab | 18 (14.1) | 0 | 18 (14.6) |
| FOLFIRI + Panitumumab | 14 (10.9) | 0 | 14 (11.4) |
| FOLFIRI | 4 (3.1) | 0 | 4 (3.3) |
| FOLFOX + Bevacizumab | 3 (2.3) | 1 (20.0) | 2 (1.6) |
| FOLFOX + Cetuximab | 1 (0.8) | 0 | 1 (0.8) |
| FOLFOX + Panitumumab | 3 (2.3) | 0 | 3 (2.4) |
| FOLFOXIRI + Bevacizumab | 2 (1.6) | 0 | 2 (1.6) |
| FOLFOXIRI | 1 (0.8) | 1 (20.0) | 0 |
| Capecitabine or UFUR | 4 (3.1) | 0 | 4 (3.3) |
| Palliative RT alone | 1 (0.8) | 0 | 1 (0.8) |
| Cetuximab + Dabrafenib + Trametinib | 2 (1.6) | 0 | 2 (1.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-H.; Lin, P.-C.; Su, W.-C.; Chan, R.-H.; Chen, P.-C.; Lin, B.-W.; Shen, M.-R.; Chen, S.-H.; Yeh, Y.-M. Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer. Diagnostics 2021, 11, 2308. https://doi.org/10.3390/diagnostics11122308
Huang Y-H, Lin P-C, Su W-C, Chan R-H, Chen P-C, Lin B-W, Shen M-R, Chen S-H, Yeh Y-M. Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer. Diagnostics. 2021; 11(12):2308. https://doi.org/10.3390/diagnostics11122308
Chicago/Turabian StyleHuang, Yi-Hsuan, Peng-Chan Lin, Wu-Chou Su, Ren-Hao Chan, Po-Chuan Chen, Bo-Wen Lin, Meng-Ru Shen, Shang-Hung Chen, and Yu-Min Yeh. 2021. "Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer" Diagnostics 11, no. 12: 2308. https://doi.org/10.3390/diagnostics11122308
APA StyleHuang, Y.-H., Lin, P.-C., Su, W.-C., Chan, R.-H., Chen, P.-C., Lin, B.-W., Shen, M.-R., Chen, S.-H., & Yeh, Y.-M. (2021). Association between Altered Oncogenic Signaling Pathways and Overall Survival of Patients with Metastatic Colorectal Cancer. Diagnostics, 11(12), 2308. https://doi.org/10.3390/diagnostics11122308






