Evaluation of MicroScan Bacterial Identification Panels for Low-Resource Settings
Abstract
:1. Introduction
Bacterial Identification in Low-Resource Settings
2. Materials and Methods
2.1. Identification Panels
2.2. Clinical Isolates and Reference Strains
2.3. Reference Identification of Clinical Isolates
2.4. Preparation of Suspension
2.5. Inoculation of Panels
2.6. Reading of the Panels
2.7. Robustness Testing
2.8. Comparison between Automated and Visual Reading, Repeatability and Inter-Observer Agreement
2.9. Definitions of Correct and Incorrect Identifications
2.10. Management of Incorrect Identifications
2.11. Ease of Use
3. Results
3.1. Results for Species Included in the MicroScan Database
3.2. Isolates Not Represented in the Database
3.3. Robustness Testing
3.4. Comparison between Automated and Visual Reading, Repeatability and Inter-Observer Agreement
3.5. Ease of Use
3.6. Stability
4. Discussion
4.1. Performance of the MicroScan Panels
4.2. Testing Isolates Not Represented in the Database, Robustness Testing
4.3. Comparison between Automated and Visual Reading, Repeatability and Inter-Observer Agreement
4.4. Adaptation to LRS: Stability, Ease-of-Use
4.5. Recommendations for Use and Further Development of the MicroScan System
4.6. Strengths and Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ombelet, S.; Ronat, J.B.; Walsh, T.; Yansouni, C.P.; Cox, J.; Vlieghe, E.; Martiny, D.; Semret, M.; Vandenberg, O.; Jacobs, J.; et al. Clinical bacteriology in low-resource settings: Today’s solutions. Lancet Infect. Dis. 2018, 18, e248–e258. [Google Scholar] [CrossRef]
- Petti, C.A.; Polage, C.R.; Quinn, T.C.; Ronald, A.R.; Sande, M.A. Laboratory Medicine in Africa: A Barrier to Effective Health Care. Clin. Infect. Dis. 2006, 42, 377–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.eucast.org (accessed on 18 February 2021).
- CLSI. M100: Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2020. [Google Scholar]
- Jacobs, J.; Hardy, L.; Semret, M.; Lunguya, O.; Phe, T.; Affolabi, D.; Yansouni, C.; Vandenberg, O. Diagnostic Bacteriology in District Hospitals in Sub-Saharan Africa: At the Forefront of the Containment of Antimicrobial Resistance. Front. Med. 2019, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.; Kaushik, K.S.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in chemical and biological methods to identify microorganisms—From past to present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [Green Version]
- Leber, A.L. Clinical Microbiology Procedures Handbook; ASM Press: Washington, DC, USA, 2016. [Google Scholar]
- Jorgensen, J.H. Manual of Clinical Microbiology, 11th ed.; Jorgensen, J.H., Carroll, K.C., Funke, G., Pfaller, M.A., Eds.; ASM Press: Washington, DC, USA, 2015; ISBN 9781555817374. [Google Scholar]
- Sandle, T. Microbial identification. In Pharmaceutical Microbiology; Elsevier Inc.: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Natale, A.; Ronat, J.-B.; Mazoyer, A.; Rochard, A.; Boillot, B.; Hubert, J.; Baillet, B.; Ducasse, M.; Mantelet, F.; Oueslati, S.; et al. The Mini-Lab: Accessible clinical bacteriology for low-resource settings. Lancet Microbe 2020, 1, 56–58. [Google Scholar] [CrossRef]
- Russell, F.M.; Biribo, S.S.N.; Selvaraj, G.; Oppedisano, F.; Warren, S.; Seduadua, A.; Mulholland, E.K.; Carapetis, J.R. As a bacterial culture medium, citrated sheep blood agar is a practical alternative to citrated human blood agar in laboratories of developing countries. J. Clin. Microbiol. 2006, 44, 3346–3351. [Google Scholar] [CrossRef] [Green Version]
- Zomorodian, K.; Javad, M.; Safaei, A.; Bazargani, A. Analysis of beta-hemolysis in human blood agars by Streptococcus pyogenes Analysis of beta-hemolysis in human blood agars by Streptococcus pyogenes. J. Microbiol. Methods 2011, 85, 233–234. [Google Scholar] [CrossRef] [PubMed]
- WHO. Technical Guidance Series (TGS) for WHO Prequalification—Diagnostic Assessment Guidance on Test Guidance on Test Method Validation for In Vitro Diagnostic Medical Devices TGS–4; Tech. Guid. Ser. WHO Prequalification—Diagnostic Assess; WHO: Geneva, Switzerland, 2017; pp. 1–23. Available online: http://apps.who.int/bookorders.%0Ahttp://apps.who.int/iris/bitstream/10665/258971/1/WHO-EMP-RHT-PQT-TGS4-2017.04-eng.pdf?ua=1 (accessed on 18 February 2021).
- Jin, W.Y.; Jang, S.J.; Lee, M.J.; Park, G.; Kim, M.J.; Kook, J.K.; Kim, D.M.; Moon, D.S.; Park, Y.J. Evaluation of VITEK 2, MicroScan, and Phoenix for identification of clinical isolates and reference strains. Diagn. Microbiol. Infect. Dis. 2011, 70, 442–447. [Google Scholar] [CrossRef]
- Rhoads, S.; Marinelli, L.; Imperatrice, C.A.; Nachamkin, I. Comparison of MicroScan WalkAway system and Vitek system for identification of gram-negative bacteria. J. Clin. Microbiol. 1995, 33, 3044–3046. [Google Scholar] [CrossRef] [Green Version]
- Snyder, J.W.; Munier, G.K.; Johnson, C.L. Direct comparison of the BD phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 2008, 46, 2327–2333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatzigeorgiou, K.S.; Sergentanis, T.N.; Tsiodras, S.; Hamodrakas, S.J.; Bagos, P.G. Phoenix 100 versus Vitek 2 in the identification of gram-positive and gram-negative bacteria: A comprehensive meta-analysis. J. Clin. Microbiol. 2011, 49, 3284–3291. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, C.M. Manual and Automated Instrumentation for Identication of Enterobacteriaceae and Other Aerobic Gram-Negative Bacilli. Clin Microbiol Rev 2005, 18, 147–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Baum, H.; Klemme, F.R.; Geiss, H.K.; Sonntag, H.G. Comparative evaluation of a commercial system for identification of gram-positive cocci. Eur. J. Clin. Microbiol. Infect. Dis. 1998, 17, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Jesumirhewe, C.; Ogunlowo, P.O.; Olley, M.; Springer, B.; Allerberger, F.; Ruppitsch, W. Accuracy of conventional identification methods used for Enterobacteriaceae isolates in three Nigerian hospitals. PeerJ 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, E.J. Rapid Identification of Bacteria and Yeast: Summary of a National Committee for Clinical Laboratory Standards Proposed Guideline. Clin. Infect. Dis. 2001, 33, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Baron, E.J.; York, M.K.; Ferraro, M.J.; Rex, J.H.; Body, B.A.; Forbes, B.A.; Poole, F.M.; Sahm, D.F.; Tenover, F.C.; Turnidge, J.D.; et al. M35-A2 Abbreviated Identification of Bacteria and, 2nd ed.; CLSI: Wayne, NJ, USA, 2008; ISBN 1562386816. [Google Scholar]
- Rand, K.H.; Tillan, M. Errors in interpretation of gram stains from positive blood cultures. Am. J. Clin. Pathol. 2006, 126, 686–690. [Google Scholar] [CrossRef] [Green Version]
- Samuel, L.P.; Balada-Llasat, J.M.; Harrington, A.; Cavagnolo, R. Multicenter assessment of gram stain error rates. J. Clin. Microbiol. 2016, 54, 1442–1447. [Google Scholar] [CrossRef] [Green Version]
- Munson, E.; Block, T.; Basile, J.; Hryciuk, J.E.; Schell, R.F. Mechanisms to assess Gram stain interpretation proficiency of technologists at satellite laboratories. J. Clin. Microbiol. 2007, 45, 3754–3758. [Google Scholar] [CrossRef] [Green Version]
- Chandler, L. Challenges in Clinical Microbiology Testing, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; ISBN 9780124157835.
- Yuan, S.; Astion, M.L.; Schapiro, J.; Limaye, A.P. Clinical impact associated with corrected results in clinical microbiology testing. J. Clin. Microbiol. 2005, 43, 2188–2193. [Google Scholar] [CrossRef] [Green Version]
- Ellner, P.D.; Myers, D.A. Preliminary evaluation of the autoSCAN-3, an instrument for automated reading and interpretation of microdilution trays: Identification of aerobic gram-negative bacilli. J. Clin. Microbiol. 1981, 14, 326–328. [Google Scholar] [CrossRef] [Green Version]
- Rhoden, D.L.; Smith, P.B.; Baker, C.N.; Schable, B. autoSCAN-4 system for identification of gram-negative bacilli. J. Clin. Microbiol. 1985, 22, 915–918. [Google Scholar] [CrossRef] [Green Version]
- Gavini, F.; Husson, M.O.; Izard, D.; Bernigaud, A.; Quiviger, B. Evaluation of Autoscan-4 for identification of members of the family Enterobacteriaceae. J. Clin. Microbiol. 1988, 26, 1586–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization—R&D Blueprint. COVID-19 Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic v.0.1; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Gillet, P.; Maltha, J.; Hermans, V.; Ravinetto, R.; Bruggeman, C.; Jacobs, J. Malaria rapid diagnostic kits: Quality of packaging, design and labelling of boxes and components and readability and accuracy of information inserts. Malar. J. 2011, 10, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, E.A.; Shaw, A.V.; Crump, J.A. Community-acquired bloodstream infections in Africa: A systematic review and meta-analysis. Lancet Infect. Dis. 2010, 10, 417–432. [Google Scholar] [CrossRef] [Green Version]
- Deen, J.; von Seidlein, L.; Andersen, F.; Elle, N.; White, N.J.; Lubell, Y. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: A systematic review. Lancet Infect. Dis. 2012, 12, 480–487. [Google Scholar] [CrossRef]
- WHO. Technical Specification Series. 2020. Available online: https://www.who.int/diagnostics_laboratory/guidance/technical-specifications-series/en/ (accessed on 18 February 2021).
- Gerth-Guyette, E.; Malacad, C.C.; Demonteverde, M.P.; Faulx, D.; Lochhead, M.J.; Lupisan, S.P.; Leader, B.T.; Tallo, V.L. Understanding user requirements to improve adoption of influenza diagnostics in clinical care within Metro Manila. Health Sci. Rep. 2018, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]


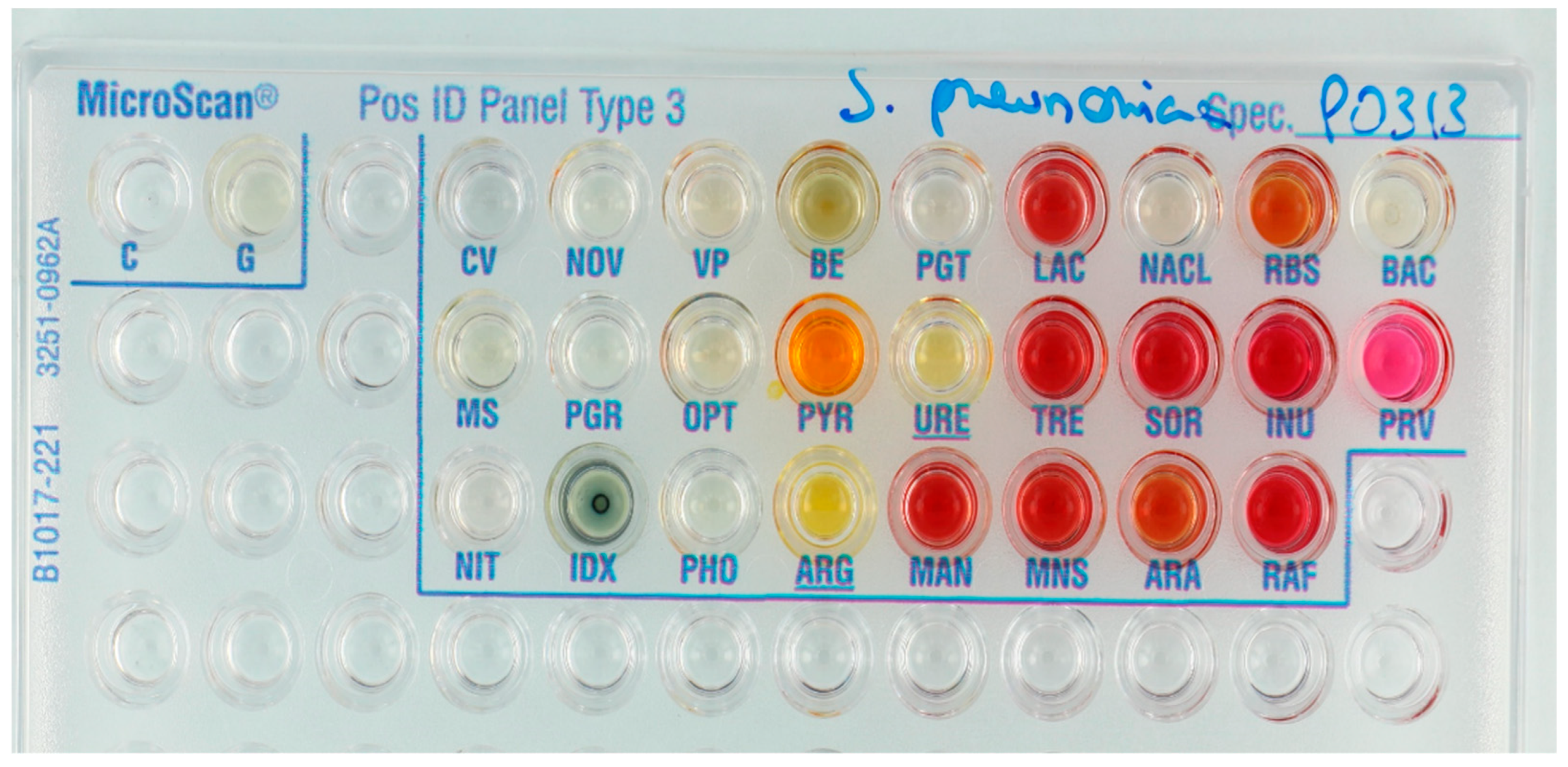
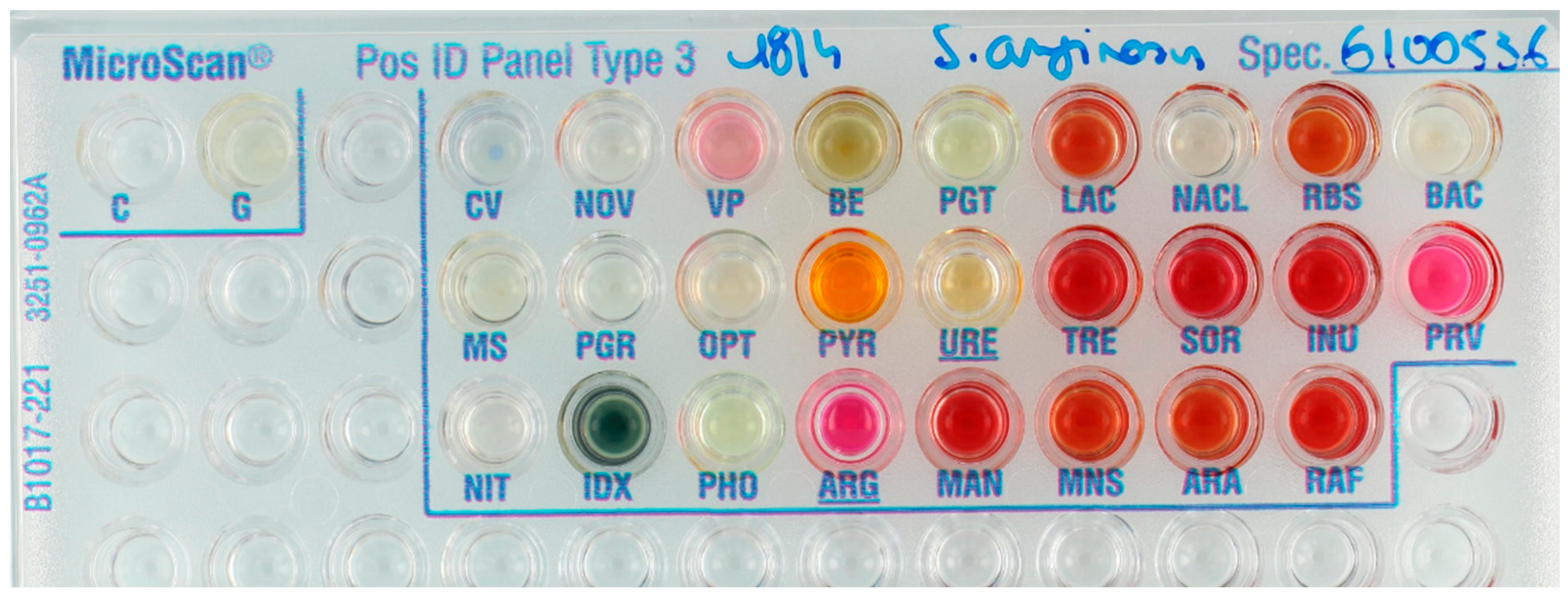
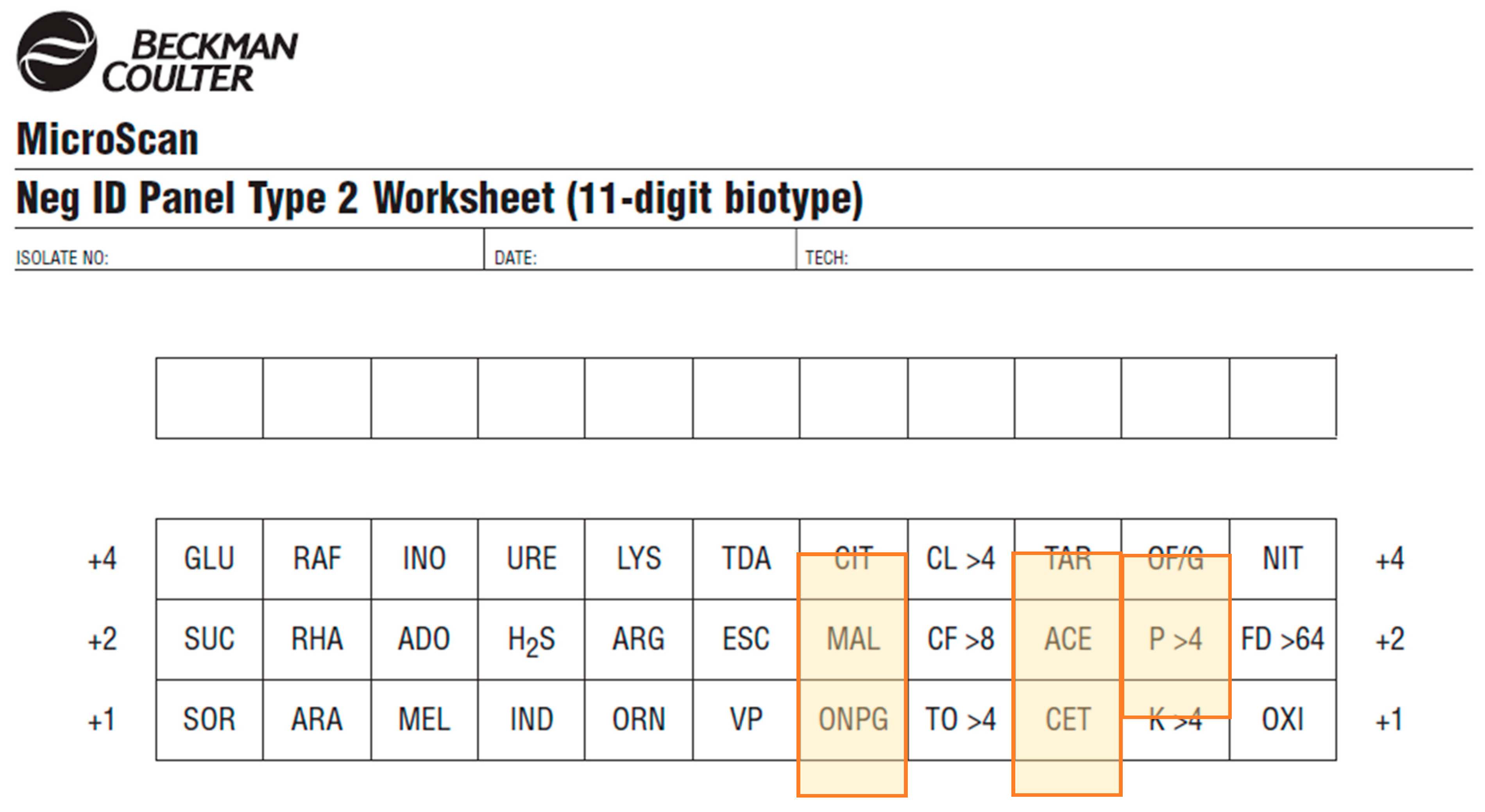
| Substrate. | Abbreviation | Panel | Substrate | Abbreviation | Panel |
|---|---|---|---|---|---|
| Glucose | GLU | Gram-negative | Lactose | LAC | Gram-positive |
| Sucrose | SUC | Gram-negative | Trehalose | TRE | Gram-positive |
| Inositol | INO | Gram-negative | Mannose | MNS | Gram-positive |
| Adonitol | ADO | Gram-negative | Ribose | RBS | Gram-positive |
| Rhamnose | RHA | Gram-negative | Inulin | INU | Gram-positive |
| Melibiose | MEL | Gram-negative | Mannitol | MAN | Gram-positive |
| Penicillin G 4 µg/mL | P4 | Gram-negative | PNP-β-d-Glucuronide | PGR | Gram-positive |
| Kanamycin 4 µg/mL | K4 | Gram-negative | PNP-β-d-Galactopyranoside | PGT | Gram-positive |
| Colistin 4 µg/mL | Cl4 | Gram-negative | Indoxyl Phosphatase | IDX | Gram-positive |
| Cephalothin 8 µg/mL | Cf8 | Gram-negative | Phosphatase | PHO | Gram-positive |
| Nitrofurantoin 64 g/mL | Fd64 | Gram-negative | Pyrrolidonyl-B-naphtylamide | PYR | Gram-positive |
| Tobramycin 4 g/mL | To4 | Gram-negative | Bile-Esculin | BE | Gram-positive |
| Cetrimide | CET | Gram-negative | Pyruvate | PRV | Gram-positive |
| Lysine | LYS | Gram-negative | Bacitracin | BAC | Gram-positive |
| Ornithine | ORN | Gram-negative | Crystal Violet | CV | Gram-positive |
| Tryptophan Deaminase | TDA | Gram-negative | Bacitracin 0.05 g/mL | MS | Gram-positive |
| Esculin | ESC | Gram-negative | Novobiocin 1.6 g/mL | NOV | Gram-positive |
| o-Nitrophenyl-d-Galactopyranoside | ONPG | Gram-negative | Optochin | OPT | Gram-positive |
| Citrate | CIT | Gram-negative | 6.5% NaCl | NACL | Gram-positive |
| Malonate | MAL | Gram-negative | Sorbitol | SOR | Both panels |
| Acetamide | ACE | Gram-negative | Raffinose | RAF | Both panels |
| Tartrate | TAR | Gram-negative | Arabinose | ARA | Both panels |
| Oxidation Base Control | OF/B | Gram-negative | Arginine | ARG | Both panels |
| Oxidation of glucose | OF/G | Gram-negative | Nitrate | NIT | Both panels |
| Hydrogen Sulfide | H2S | Gram-negative | Voges-Proskauer | VP | Both panels |
| Indole | IND | Gram-negative | Urea | URE | Both panels |
| Decarboxylase Base Control | DCB | Gram-negative |
| Species | N | Correct Identification | Incorrect Identification | Identification Not Possible | ||
|---|---|---|---|---|---|---|
| High Probability Score | Low Probability Score | High Probability Score | Low Probability Score | |||
| Enterobacterales | ||||||
| Escherichia coli | 28 | 24 | 3 | - | 1 | - |
| Klebsiella pneumoniae | 13 | 13 | - | - | - | - |
| Klebsiella oxytoca | 3 | 3 | - | - | - | - |
| Enterobacter cloacae complex | 15 | 14 | - | - | 1 | - |
| Citrobacter freundii complex | 9 | 9 | - | - | - | - |
| Kluyvera ascorbata | 1 | 1 | - | - | - | - |
| Salmonella Typhi | 10 | 10 | - | - | - | - |
| Salmonella Paratyphi A | 10 | 9 | - | 1 | - | - |
| Salmonella Typhimurium | 10 | 10 | - | - | - | - |
| Salmonella Choleraesuis | 9 | 8 | - | - | 1 | - |
| Shigella species | 10 | 9 | - | - | 1 | - |
| Morganella morganii | 4 | 4 | - | - | - | - |
| Proteus mirabilis | 7 | 7 | - | - | - | - |
| Providencia rettgerii | 1 | 1 | - | - | - | - |
| Enterobacterales | 130 | 122 (93.8%) | 3 (2.3%) | 1 (0.8%) | 4 (3.1%) | 0 (0%) |
| Non-fermenting Gram-negative organisms and Aeromonas/Vibrio species | ||||||
| Pseudomonas aeruginosa | 11 | 11 | - | - | - | - |
| Acinetobacter baumannii | 9 | 8 | - | 1 | - | - |
| Achromobacter xylosoxidans | 10 | 8 | - | 1 | 1 | - |
| Burkholderia cepacia | 15 | 13 | 1 | - | 1 | - |
| Stenotrophomonas maltophilia | 15 | 14 | 1 | - | - | - |
| Aeromonas species | 3 | 2 | - | - | 1 | - |
| Vibrio alginolyticus | 2 | 2 | - | - | - | - |
| Non-fermenters | 65 | 58 (89.2%) | 2 (3.1%) | 2 (3.1%) | 3 (4.6%) | 0 (0%) |
| Total Gram-negative isolates | ||||||
| Total Gram-negative | 195 | 180 (92.3%) | 5 (2.6%) | 3 (1.5%) | 7 (3.6%) | - |
| Species | N | Correct Identification | Incorrect Identification | Identification Not Possible | ||
| High Probability Score | Low Probability Score | High Probability Score | Low Probability Score | |||
| Micrococcaceae | ||||||
| Staphylococcus aureus | 14 | 14 | - | - | - | - |
| Staphylococcus hominis | 3 | 2 | 1 | - | - | - |
| Staphylococcus epidermidis | 11 | 10 | - | 1 | - | - |
| Staphylococcus lugdunensis | 3 | 3 | - | - | - | - |
| Staphylococcus lentus | 1 | - | - | 1 | - | - |
| Staphylococcus haemolyticus | 6 | 3 | - | 2 | 1 | - |
| Listeria monocytogenes | 2 | 2 | - | - | - | - |
| Micrococcus species | 4 | 4 | - | - | - | - |
| Total Staphylococcus/Micrococcus | 44 | 38 (86.4%) | 1 (2.3%) | 4 (9.1%) | 1 (2.3%) | 0 (0%) |
| Streptococcaceae | ||||||
| Enterococcus faecalis | 7 | 7 | - | - | - | - |
| Enterococcus faecium | 7 | 4 | - | 2 | - | 1 |
| Enterococcus casseliflavus | 1 | - | - | 1 | - | - |
| Streptococcus agalactiae | 13 | 11 | 1 | - | 1 | - |
| Streptococcus anginosus | 8 | 2 | 2 | 3 | - | 1 |
| Streptococcus pneumoniae | 12 | 9 | - | 2 | 1 | - |
| Streptococcus gallolyticus * | 10 | 9 | 1 | - | - | - |
| Streptococcus lutetiensis * | 1 | - | - | - | 1 | - |
| Streptococcus pyogenes | 13 | 13 | - | - | - | - |
| Streptococcus dysgalactiae (group C & G) | 6 | 6 | - | - | - | - |
| Streptococcus mitis | 5 | 2 | 3 | - | - | - |
| Streptococcus salivarius | 1 | 1 | - | - | - | - |
| Total Streptococcus/Enterococcus species | 84 | 64 (76.2%) | 7 (8.3%) | 8 (9.5%) | 3 (3.6%) | 2 (2.4%) |
| Total Gram-positive isolates | ||||||
| Total Gram-positive | 128 | 102 (79.7%) | 8 (6.3%) | 12 (9.4%) | 4 (3.9%) | 2 (1.6%) |
| Overall total | 323 | 282 (87.3%) | 13 (4.0%) | 15 (4.6%) | 11 (3.4%) | 2 (0.6%) |
| Species | N | Identification Not Possible | Low Probability Identification | High Probability Identification |
|---|---|---|---|---|
| Gram-positive bacilli (Bacillus & Corynebacterium species) | 16 | 1 | 8 | 7 |
| Streptococcus suis | 11 | - | 5 | 6 |
| Burkholderia species (non-cepacia, non-pseudomallei) | 8 | 2 | 2 | 4 |
| Yeast species (on Gram-positive panel) | 9 | - | - | 9 |
| Yeast species (on Gram-negative panel) | 5 | - | 5 | - |
| Total (species not represented in MicroScan database) | 49 * | 3 (6.1%) | 20 (40.8%) | 26 (53.1%) |
| Species/Serotype * | Nr. of Isolates with Incorrect Identification/N | Incorrect Result by MicroScan | Probability Score | Clinical Relevance of Error |
|---|---|---|---|---|
| Gram-negative species | ||||
| Escherichia coli | 1/28 | Escherichia fergusonii | 67.88% | Limited |
| Enterobacter cloacae | 1/15 | Enterobacter amnigenus | 58.26% | Limited |
| Salmonella Paratyphi A | 1/10 | Salmonella Typhi | 98.87% | Moderate: pathogens are clinically similar, but distinction has epidemiological relevance |
| Salmonella Choleraesuis | 1/9 | Salmonella Typhi | 57.06% | High: pathogens are both clinically and epidemiologically distinct |
| Shigella species | 1/10 | Klebsiella ozaenae | 72.69% | High: pathogens have different clinical presentations; Shigella infection is of high epidemiological importance (outbreak potential) |
| Acinetobacter baumannii | 1/9 | Acinetobacter lwoffii | 92.88% | Moderate: pathogens may have different clinical presentation |
| Achromobacter xylosoxidans | 2/10 | Rhizobactrum radiobacter Burkholderia cepacia | 49.88% 94.19% | Limited |
| Aeromonas caviae | 1/3 | Aeromonas hydrophila | 61.71% | Limited |
| Gram-positive species | ||||
| Staphylococcus epidermidis | 1/11 | Staphylococcus hyicus | 91.14% | Limited |
| Staphylococcus lentus | 1/1 | Staphylococcus sciuri | 96.73% | Limited |
| Staphylococcus haemolyticus | 3/6 | Staphylococcus lugdunensis Staphylococcus lugdunensis | 53.67% 93.66% | Moderate: pathogens may have different clinical presentation |
| Enterococcus faecium | 2/7 | Enterococcus raffinosus Enterococcus durans | 95.39% 99.68% | Moderate: pathogens have different clinical significance |
| Enterococcus casseliflavus | 1/1 | Enterococcus gallinarum | 59.68% | Limited: both are associated with increased vancomycin resistance |
| Streptococcus anginosus | 3/8 | Streptococcus mitis/oralis Streptococcus salivarius Streptococcus parasanguinis | 86.99% 88.56% 99.99% | High: Streptococcus anginosus is associated with purulent infections; other viridans streptococci are more likely contaminants or implicated in infectious endocarditis |
| Streptococcus pneumoniae | 3/12 | Rhodococcus equi Gemella species Gemella species | 99.99% 51.48% 95.79% | High: Streptococcus pneumoniae is a more common pathogen than Rhodococcus equi or Gemella and clinical presentation is very different |
| Streptococcus agalactiae | 1/13 | Streptococcus dysgalactiae | 99.99% | Limited; except for screening in pregnancy |
| Species. | Algorithm Used (for Gram-Positive Panels) | N | Number of Isolates with this Result | Result |
|---|---|---|---|---|
| Species not represented in the database | ||||
| Yeast species | Micrococcus | 9 | 9 | Staphylococcus cohnii |
| Streptococcus | 9 | Rhodococcus equi | ||
| Corynebacterium species | Micrococcus | 6 | 1 | Micrococcus species |
| Streptococcus | 6 | 1 | Rhodococcus equi | |
| Bacillus species | Micrococcus | 10 | 4 | Staphylococcus auricularis |
| Gram-positive species on Gram-negative panel | ||||
| Streptococcus dysgalactiae | 2 | 2 | Providencia rustigiannii | |
| Streptococcus suis | 10 | 9 | Providencia rustigiannii | |
| Staphylococcus lentus | 1 | 1 | Providencia rustigiannii | |
| Streptococcus agalactiae | 5 | 1 | Providencia rustigiannii | |
| Gram-negative species on Gram-positive panel | ||||
| Escherichia coli/paracoli | Streptococcus | 10 | 3 | Rhodococcus equi |
| Acinetobacter baumannii | Micrococcus | 4 | 3 | Staphylococcus cohnii |
| Micrococcus | 1 | Staphylococcus auricularis | ||
| Streptococcus | 1 | Rhodococcus equi | ||
| Stenotrophomonas maltophilia | Streptococcus | 4 | 2 | Rhodococcus equi |
| Burkholderia cepacia | Streptococcus | 4 | 3 | Rhodococcus equi |
| Burkholderia thailandensis | Streptococcus | 4 | 1 | Rhodococcus equi |
| Burkholderia ubonensis | Streptococcus | 1 | 1 | Rhodococcus equi |
| Gram-positive species using incorrect algorithm on Gram-positive panel | ||||
| Staphylococcus aureus | Streptococcus | 14 | 3 | Rhodococcus equi |
| Micrococcus species | Streptococcus | 4 | 2 | Rhodococcus equi |
| Staphylococcus lentus | Streptococcus | 1 | 1 | Rhodococcus equi |
| Streptococcus gallolyticus | Micrococcus | 10 | 9 | Listeria monocytogenes |
| Streptococcus pneumoniae | Micrococcus | 12 | 9 | Micrococcus species |
| 2 | Staphylococcus auricularis | |||
| Streptococcus pyogenes | Micrococcus | 13 | 2 | Staphylococcus auricularis |
| Streptococcus agalactiae | Micrococcus | 13 | 1 | Staphylococcus auricularis |
| Streptococcus anginosus | Micrococcus | 8 | 1 | Staphylococcus auricularis |
| Streptococcus mitis/oralis | Micrococcus | 5 | 1 | Staphylococcus auricularis |
| Species. | N | Panel/Algorithm Used | Number “Identification not Possible” (%) | Number Low Probability Identification (%) | Number High Probability Identification (%) |
|---|---|---|---|---|---|
| Gram-negative species tested on Gram-positive panels and vice versa | |||||
| Enterobacterales | 41 | Gram-positive | 27 | 9 | 5 |
| Non-fermenters | 23 | 10 | 6 | 7 | |
| Total Gram-negatives on Gram-positive panel | 64 | 37 (57.8%) | 15 (23.4%) | 12 (18.8%) | |
| Staphylococcus species | 16 | Gram-negative | 1 | 11 | 4 |
| Streptococcus species | 53 | 15 | 12 | 26 | |
| Gram-positive bacilli (Bacillus & Corynebacterium species) | 11 | 2 | 4 | 5 | |
| Total Gram-positive on Gram-negative panel | 80 | 18 (22.5%) | 27 (33.8%) | 35 (43.8%) | |
| Staphylococcus/Micrococcus species using Streptococcus species algorithm and vice versa | |||||
| Staphylococcus/Micrococcus species and Gram-positive rods | 60 | Streptococcus algorithm | 35 | 11 | 14 |
| Streptococcus/Enterococcus species | 95 | Micrococcus algorithm | 27 | 24 | 44 |
| Total Gram-positive with incorrect algorithm | 155 | 62 (40.0%) | 35 (22.6%) | 58 (37.4%) | |
| Total tested on incorrect panel/incorrect algorithm | 299 | 117 (39.1%) | 77 (25.8%) | 105 (35.1%) | |
| Favorable Observations | Unfavorable Observations |
|---|---|
| Storage of panels and reagents | |
| Long shelf life of the panels and of most reagents | VP2 reagent must be reconstituted with 95% ethanol (not provided); after reconstitution, vial is stable for only 2 weeks |
| Panels and some reagents can be stored at room temperature | Many reagents (nitrate, VP, ferric chloride, Kovac’s) require storage at 2–8 °C |
| Instructions for use, labeling, and packaging | |
| Panels are packaged separately with desiccant, (StripPax®, Multisorb Technologies, New York, NY, USA) | Reconstitution of VP2 reagent not explained in IFU, only on box of reagent |
| Sturdy outer packaging | |
| IFU were delivered at request by the distributor | IFU are not delivered in the box with the products; they are available online but not easy to find |
| Flesch Kincaid Grade level of IFU was 9.14 for the Gram-negative IFU, 11.06 for the Gram-positive IFU and 10.30 for the MSF panel IFU; these levels point to a high level of education needed to comprehend the text | |
| Instructions in IFU for interpretation of negative/positive wells are often ambiguous.e.g., yellow to orange color for carbohydrate tests must be considered positive, whereas orange to red must be considered negative | |
| Reagent bottles are labeled with the test name: easy to add right reagent to right test well | Use of reagent names is inconsistent between IFU and reagent bottles (e.g., 5% alpha naphtol in IFU, VP2 on bottle) |
| The PYR test reagent has “PEP” on the bottle and not “PYR” | |
| Limited space for writing patient identification and sample number on the panel | |
| Inoculation and reading | |
| Most panels could be inoculated using the Prompt™ system | Prompt™ is not advised for use in slow-growing Streptococcus species; we repeated testing of 14 Streptococcus isolates with turbidity standard method due to incorrect results with Prompt™; 50% of these had a correct result with turbidity standard method |
| Prompt ™ was easy to use and time-efficient | |
| RENOK system was easy to use and time-efficient | Large amounts of plastic waste associated with the RENOK system |
| Difficulties in disposing of waste without spillage | |
| Difficulties in incubation of panels without spillage | |
| Favorable observations | Unfavorable observations |
| IFU recognizes that interpretation of test results requires trained clinical staff | Visual reading of panels was considered difficult as positivity of test wells was often doubtful |
| Automated reading was more user-friendly as it removed subjectivity and eliminated clerical errors | autoSCAN-4 does not indicate automatically that panels must be re-incubated |
| autoSCAN-4 reader dimensions are very large: 48.5 cm × 24 cm × 58 cm | |
| Prototype microplate viewer (JP Selecta) with magnifier and interchangeable white/black background considerably facilitated the reading | Both white and black background needed for reading of wells |
| Reagents can be added only after visual assessment of certain wells indicating the need for re-incubation; panels must be re-incubated if specific combinations of wells are negative—there are 6 of these combinations that consist each of 1 to 3 wells unevenly distributed on the panels | |
| Different reading times for different reagents | |
| Re-incubation was required for 15.3% of panels in our study; 13.3% (27/203) of Gram-negative isolates (mostly non-fermenters) and 17.7% (29/164) of Gram-positive isolates (mainly Streptococcus isolates). | |
| Software and Biotype Lookup Tool | |
| LabPro software and reader easy to use | Beckman Coulter worksheets not delivered with the panels; available online but hard to find |
| Calibration and QC of reader are alerted by the reader automatically and do not require user input | Test position on worksheet different from test position on the panel (for Gram-negative panel) (Figure 5). |
| Biotype Lookup Tools freely available online | |
| Both the online Biotype Lookup Tool and LabPro software propose additional tests to confirm low-probability identification | Many of the confirmatory tests proposed by the software are unlikely to be easily available in LRS Examples of recommended confirmatory tests:
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ombelet, S.; Natale, A.; Ronat, J.-B.; Vandenberg, O.; Hardy, L.; Jacobs, J. Evaluation of MicroScan Bacterial Identification Panels for Low-Resource Settings. Diagnostics 2021, 11, 349. https://doi.org/10.3390/diagnostics11020349
Ombelet S, Natale A, Ronat J-B, Vandenberg O, Hardy L, Jacobs J. Evaluation of MicroScan Bacterial Identification Panels for Low-Resource Settings. Diagnostics. 2021; 11(2):349. https://doi.org/10.3390/diagnostics11020349
Chicago/Turabian StyleOmbelet, Sien, Alessandra Natale, Jean-Baptiste Ronat, Olivier Vandenberg, Liselotte Hardy, and Jan Jacobs. 2021. "Evaluation of MicroScan Bacterial Identification Panels for Low-Resource Settings" Diagnostics 11, no. 2: 349. https://doi.org/10.3390/diagnostics11020349
APA StyleOmbelet, S., Natale, A., Ronat, J.-B., Vandenberg, O., Hardy, L., & Jacobs, J. (2021). Evaluation of MicroScan Bacterial Identification Panels for Low-Resource Settings. Diagnostics, 11(2), 349. https://doi.org/10.3390/diagnostics11020349







