Data Integration Reveals the Potential Biomarkers of Circulating MicroRNAs in Osteoarthritis
Abstract
1. Introduction
2. Brief Introduction to miRNAs and c-miRNAs
3. c-miRNA Signature as Potential Biomarker for OA Diagnosis
4. Therapeutic Potential of c-miRNAs in OA
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAMTS5 | A disintegrin-like and metalloproteinase with thrombospondin-1 motifs 5 |
| AGO | Argonaute protein |
| AKT | Serine/threonine-specific protein kinase |
| ATG14 | Autophagy related 14 |
| Bcl-2 | B-cell lymphoma 2 |
| BECN1 | Beclin 1 |
| BMI | Body mass index |
| C1, C2, C2C | Types I and II collagen marker degradation markers |
| Camk2d | Calcium/calmodulin-dependent protein kinase type II delta |
| CASPASE-3 | Cysteine-dependent aspartate-directed proteases 3 |
| CBFB | Core-binding factor subunit beta |
| c-miRNA | Circulating microRNA |
| Col2α1 | The type II collagen gene |
| Coll2-1 NO2 | The nitrated form of α-helical region of type II collagen |
| COMP | Oligomeric matrix protein |
| COX2 | Prostaglandin-endoperoxide synthase 2 |
| CPII | C-propeptide of type II collagen |
| CS846 | Chondroitin sulphate 846 |
| CTXI | C-terminal cross-linked telopeptide of type I collagen |
| CTXI alpha and CTXI beta | Alpha and beta isomerised versions of the CTXI |
| CTXII | C-terminal crosslinked telopeptide of type II collagen |
| DMM | Medical meniscus |
| ERG | Erythroblastosis virus E26 oncogene homolog-related gene |
| EZH2 | Enhancer of zeste homolog 2 |
| FGF2 | Fibroblast growth factor 2 |
| FGFR | Fibroblast growth factor receptor |
| HA | Hyaluronic acid |
| HADC2 | Histone deacetylase 2 |
| HDAC4 | Histone deacetylase 4 |
| HDL | High-density lipoprotein |
| HIF-1α | Hypoxia-inducible factor 1-alpha |
| HIF-2α | Hypoxia-inducible factor 2-alpha |
| IGFBP5 | Insulin-like growth factor-binding protein 5 |
| IGFIR | Insulin-like growth factor 1 receptor |
| IGFR | Insulin-like growth factor 1 receptor |
| IL | Interleukine |
| iNOS | Inducible nitric oxide synthase |
| InsR | Insulin receptor |
| IRAK1 | Interleukin-1 receptor-associated kinase 1 |
| LNA-ASO | Locked nucleic acid antisense oligonucleotide |
| LncRNA H19 | Long non-coding RNA H19 |
| MAP1LC3 | Microtubule-associated proteins 1A/1B light chain 3B |
| miRNA | microRNA |
| MMP-13 | Matrix metalloproteinase 13 |
| MMP3 | Matrix metalloproteinase-3 |
| NLRP3 | The Nacht, leucine-rich repeat and pyrin domain containing protein 3 |
| NTXI | The cross-linked N-telopeptide of type I collagen |
| OA | osteoarthritis |
| p16INK4α | Cyclin dependent kinase inhibitor 2A |
| PARP p85 | Poly (ADP-ribose) polymerase p85 |
| PI3K | Phosphoinositide 3-kinases |
| PIIANP | N-propeptide of collagen IIA |
| Ppp3r2 | Protein phosphatase 3 regulatory subunit B |
| PTEN | Phosphatase and tensin homolog |
| qPCR | Quantitative real-time PCR |
| RALA | Ras-related protein Ral-A |
| REGγ | Langerhans regenerating protein γ |
| RUNX2 | Runt-related transcription factor 2 |
| SHIP1 | Src homology 2 domain containing inositol polyphosphate 5-phosphatase 1 |
| SIRT1 | Sirtuin 1 |
| Smad2 | Mothers against decapentaplegic homolog 2 |
| Smad3 | Mothers against decapentaplegic homolog 3 |
| Smad4 | SMAD family member 4 |
| SPP1 | Secreted phosphoprotein 1 |
| TGF | Transforming growth factor |
| TGIF2 | TGFB-induced factor homeobox 2 |
| TIMP2 | Tissue inhibitor of metalloproteinases 2 |
| TRAF3 | TNF receptor-associated factor 3 |
| TRAF6 | TNF receptor associated factor 6 |
| ULK1 | Unc-51-like autophagy activating kinase 1 |
| VEGF | Vascular endothelial growth factor |
| VEGFA | Vascular endothelial growth factor A |
| WISP1 | Wnt1-inducible signaling pathway protein 1 |
References
- Beyer, C.; Zampetaki, A.; Lin, N.Y.; Kleyer, A.; Perricone, C.; Iagnocco, A.; Distler, A.; Langley, S.R.; Gelse, K.; Sesselmann, S.; et al. Signature of circulating microRNAs in osteoarthritis. Ann. Rheum. Dis. 2015, 74, e18. [Google Scholar] [CrossRef]
- Chen, D.; Shen, J.; Zhao, W.; Wang, T.; Han, L.; Hamilton, J.L.; Im, H.J. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 2017, 5, 16044. [Google Scholar] [CrossRef]
- Panagopoulos, P.; Lambrou, G. The Involvement of MicroRNAs in Osteoarthritis and Recent Developments: A Narrative Review. Mediterr. J. Rheumatol. 2018, 29, 67–79. [Google Scholar] [CrossRef]
- OECD. Health at a Glance 2019. In Health at a Glance; OECD: Paris, France, 2019; ISBN 9789264382084. [Google Scholar]
- Reginster, J.Y. The prevalence and burden of arthritis. Rheumatology 2002, 41 (Suppl. S1), 3–6. [Google Scholar] [CrossRef] [PubMed]
- Bitton, R. The economic burden of osteoarthritis. Am. J. Manag. Care 2009, 15, S230–S235. [Google Scholar] [PubMed]
- Michael, J.W.P.; Schlüter-Brust, K.U.; Eysel, P. The Epidemiology, Etiology, Diagnosis, and Treatment of Osteoarthritis of the Knee. Dtsch. Aerzteblatt Online 2010, 107, 152–162. [Google Scholar] [CrossRef]
- Kwoh, C.K. Epidemiology of osteoarthritis. Epidemiol. Aging 2012, 26, 523–536. [Google Scholar] [CrossRef]
- Li, Q.; Amano, K.; Link, T.M.; Ma, C.B. Advanced Imaging in Osteoarthritis. Sports Health Multidiscip. Approach 2016, 8, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Litwic, A.; Edwards, M.H.; Dennison, E.M.; Cooper, C. Epidemiology and burden of osteoarthritis. Br. Med. Bull. 2013, 105, 185–199. [Google Scholar] [CrossRef]
- Driban, J.B.; Harkey, M.S.; Liu, S.H.; Salzler, M.; McAlindon, T.E. Osteoarthritis and Aging: Young Adults with Osteoarthritis. Curr. Epidemiol. Rep. 2020, 7, 9–15. [Google Scholar] [CrossRef]
- Hame, S.L.; Alexander, R.A. Knee osteoarthritis in women. Curr. Rev. Musculoskelet. Med. 2013, 6, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Plotnikoff, R.; Karunamuni, N.; Lytvyak, E.; Penfold, C.; Schopflocher, D.; Imayama, I.; Johnson, S.T.; Raine, K. Osteoarthritis prevalence and modifiable factors: A population study. BMC Public Health 2015, 15, 1195. [Google Scholar] [CrossRef]
- Vina, E.R.; Kwoh, C.K. Epidemiology of osteoarthritis: Literature update. Curr. Opin. Rheumatol. 2018, 30, 160–167. [Google Scholar] [CrossRef]
- Fernández-Moreno, M.; Rego, I.; Carreira-Garcia, V.; Blanco, F.J. Genetics in osteoarthritis. Curr. Genom. 2008, 9, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Barter, M.J.; Bui, C.; Young, D.A. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthr. Cartil. 2012, 20, 339–349. [Google Scholar] [CrossRef]
- Warner, S.C.; Valdes, A.M. Genetic association studies in osteoarthritis: Is it fairytale? Curr. Opin. Rheumatol. 2017, 29, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Fathollahi, A.; Aslani, S.; Jamshidi, A.; Mahmoudi, M. Epigenetics in osteoarthritis: Novel spotlight. J. Cell. Physiol. 2019, 234, 12309–12324. [Google Scholar] [CrossRef]
- Thomas, A.C.; Hubbard-Turner, T.; Wikstrom, E.A.; Palmieri-Smith, R.M. Epidemiology of Posttraumatic Osteoarthritis. J. Athl. Train. 2017, 52, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Coggon, D.; Reading, I.; Croft, P.; McLaren, M.; Barrett, D.; Cooper, C. Knee osteoarthritis and obesity. Int. J. Obes. Relat. Metab. Disord. 2001, 25, 622–627. [Google Scholar] [CrossRef]
- Dubé, C.E.; Liu, S.H.; Driban, J.B.; McAlindon, T.E.; Eaton, C.B.; Lapane, K.L. The relationship between smoking and knee osteoarthritis in the Osteoarthritis Initiative. Osteoarthr. Cartil. 2016, 24, 465–472. [Google Scholar] [CrossRef]
- Kang, A.H.; Kim, M.R.; Shin, J.S.; Lee, J.; Lee, Y.J.; Park, Y.; Nam, D.; Kim, E.J.; Ha, I.H. Association between alcohol consumption and osteoarthritis prevalence in Korea as assessed by the alcohol use disorders identification test (AUDIT): A cross-sectional study. BMC Public Health 2020, 20, 227. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. Diet, nutrition and osteoarthritis. BMC Musculoskelet Disord. 2015, 16, S7. [Google Scholar] [CrossRef][Green Version]
- Messina, O.D.; Vidal Wilman, M.; Vidal Neira, L.F. Nutrition, osteoarthritis and cartilage metabolism. Aging Clin. Exp. Res. 2019, 31, 807–813. [Google Scholar] [CrossRef]
- Cooper, C.; Coggon, D. Physical activity and knee osteoarthritis. Lancet 1999, 353, 2177e8. [Google Scholar] [CrossRef]
- Coggon, D.; Croft, P.; Kellingray, S.; Barrett, D.; McLaren, M.; Cooper, C. Occupational physical activities and osteoarthritis of the knee. Arthritis Rheum. 2000, 7, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Verbeek, J.; Mischke, C.; Robinson, R.; Ijaz, S.; Kuijer, P.; Kievit, A.; Ojajärvi, A.; Neuvonen, K. Occupational Exposure to Knee Loading and the Risk of Osteoarthritis of the Knee: A Systematic Review and a Dose-Response Meta-Analysis. Saf. Health Work 2017, 8, 130–142. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, D.F.; Leeb, B.F.; Muthuri, S.G.; Doherty, M.; Zhang, W. Occupational risk factors for osteoarthritis of the knee: A meta-analysis. Osteoarthr. Cartil. 2011, 19, 829–839. [Google Scholar] [CrossRef]
- Yucesoy, B.; Charles, L.E.; Baker, B.; Burchfiel, C.M. Occupational and genetic risk factors for osteoarthritis: A review. Work 2015, 50, 261–273. [Google Scholar] [CrossRef]
- Liem, Y.; Judge, A.; Kirwan, J.; Ourradi, K.; Li, Y.; Sharif, M. Multivariable logistic and linear regression models for identification of clinically useful biomarkers for osteoarthritis. Sci. Rep. 2020, 10, 11328. [Google Scholar] [CrossRef] [PubMed]
- Denoble, A.E.; Huffman, K.M.; Stabler, T.V.; Kelly, S.J.; Hershfield, M.S.; McDaniel, G.E.; Coleman, R.E.; Kraus, V.B. Uric acid is a danger signal of increasing risk for osteoarthritis through inflammasome activation. Proc. Natl. Acad. Sci. USA 2011, 108, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Ishijima, M.; Kaneko, H.; Kaneko, K. The evolving role of biomarkers for osteoarthritis. Ther. Adv. Musculoskelet. Dis. 2014, 6, 144–153. [Google Scholar] [CrossRef]
- Hamilton, J.P. Epigenetics: Principles and Practice. Dig. Dis. 2011, 29, 130–135. [Google Scholar] [CrossRef]
- Kung, L.H.W.; Zaki, S.; Ravi, V.; Rowley, L.; Smith, M.M.; Bell, K.M.; Bateman, J.F.; Little, C.B. Utility of circulating serum miRNAs as biomarkers of early cartilage degeneration in animal models of post-traumatic osteoarthritis and inflammatory arthritis. Osteoarthr. Cartil. 2017, 25, 426–434. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 3, 402. [Google Scholar] [CrossRef]
- Lao, T.D.; Le, T.A.H. MicroRNAs: Biogenesis, Functions and Potential Biomarkers for Early Screening, Prognosis and Therapeutic Molecular Monitoring of Nasopharyngeal Carcinoma. Processes 2020, 8, 966. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Rapisuwon, S.; Vietsch, E.E.; Wellstein, A. Circulating biomarkers to monitor cancer progression and treatment. Comput. Struct. Biotechnol. J. 2016, 14, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Zhang, J.; Zhang, W.; Huang, R.S. Circulating MicroRNAs as Biomarkers for Inflammatory Diseases. MicroRNA 2013, 2, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Min, P.K.; Chan, S.Y. The biology of circulating microRNAs in cardiovascular disease. Eur. J. Clin. Investig. 2015, 45, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Duttagupta, R.; Jiang, R.; Gollub, J.; Getts, R.C.; Jones, K.W. Impact of Cellular miRNAs on Circulating miRNA Biomarker Signatures. PLoS ONE 2011, 6, e20769. [Google Scholar] [CrossRef]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef]
- Thuy, L.H.A.; Thuan, L.D.; Phuong, T.K. DNA Hypermethylation in Breast Cancer. In Breast Cancer—From Biology to Medicine; InTech: London, UK, 2017. [Google Scholar]
- Weng, L.; Wu, X.; Gao, H.; Mu, B.; Li, X.; Wang, J.H.; Guo, C.; Jin, J.M.; Chen, Z.; Covarrubias, M.; et al. MicroRNA profiling of clear cell renal cell carcinoma by whole-genome small RNA deep sequencing of paired frozen and formalin-fixed, paraffin-embedded tissue specimens. J. Pathol. 2010, 222, 41–51. [Google Scholar] [CrossRef]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013, 774, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.P.; Ruprecht, K.; Meese, E. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE 2009, 13, e7440. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Rosa, A.; Guedes, L.C.; Fonseca, B.V.; Gotovac, K.; Violante, S.; Mestre, T.; Coelho, M.; Rosa, M.M.; Martin, E.R.; et al. Convergence of miRNA expression profiling, α-synuclein interacton and GWAS in Parkinson’s disease. PLoS ONE 2011, 6, e25443. [Google Scholar] [CrossRef] [PubMed]
- Laczny, C.; Leidinger, P.; Haas, J.; Ludwig, N.; Backes, C.; Gerasch, A.; Kaufmann, M.; Vogel, B.; Katus, H.A.; Meder, B.; et al. miRTrail—A comprehensive webserver for analyzing gene and miRNA patterns to enhance the understanding of regulatory mechanisms in diseases. BMC Bioinform. 2012, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Leidinger, P.; Vogel, B.; Backes, C.; ElSharawy, A.; Galata, V.; Mueller, S.C.; Marquart, S.; Schrauder, M.G.; Strick, R.; et al. miRNAs can be generally associated with human pathologies as exemplified for miR-144. BMC Med. 2014, 12, 224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Soyocak, A.; Kurt, H.; Ozgen, M.; Turgut Cosan, D.; Colak, E.; Gunes, H.V. miRNA-146a, miRNA-155 and JNK expression levels in peripheral blood mononuclear cells according to grade of knee osteoarthritis. Gene 2017, 627, 207–211. [Google Scholar] [CrossRef]
- Okuhara, A.; Nakasa, T.; Shibuya, H.; Niimoto, T.; Adachi, N.; Deie, M.; Ochi, M. Changes in microRNA expression in peripheral mononuclear cells according to the progression of osteoarthritis. Mod. Rheumatol. 2012, 22, 446–457. [Google Scholar] [CrossRef]
- Skrzypa, M.; Szala, D.; Gablo, N.; Czech, J.; Pajak, J.; Kopanska, M.; Trzeciak, M.; Gargasz, K.; Snela, S.; Zawlik, I. miRNA-146a-5p is upregulated in serum and cartilage samples of patients with osteoarthritis. Pol. Przegl. Chir. 2019, 91, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, J.C.; Millet, M.; Croset, M.; Sornay-Rendu, E.; Borel, O.; Chapurlat, R. Association of circulating microRNAs with prevalent and incident knee osteoarthritis in women: The OFELY study. Arthritis Res. Ther. 2020, 22, 1–12. [Google Scholar] [CrossRef]
- Cuadra, V.M.B.; González-Huerta, N.C.; Romero-Córdoba, S.; Hidalgo-Miranda, A.; Miranda-Duarte, A. Altered expression of circulating microRNA in plasma of patients with primary osteoarthritis and in silico analysis of their pathways. PLoS ONE 2014, 9, e97690. [Google Scholar] [CrossRef]
- Li, Y.-H.; Tavallaee, G.; Tokar, T.; Nakamura, A.; Sundararajan, K.; Weston, A.; Sharma, A.; Mahomed, N.N.; Gandhi, R.; Jurisica, I.; et al. Identification of synovial fluid microRNA signature in knee osteoarthritis: Differentiating early- and late-stage knee osteoarthritis. Osteoarthr. Cartil. 2016, 24, 1577–1586. [Google Scholar] [CrossRef]
- Ntoumou, E.; Tzetis, M.; Braoudaki, M.; Lambrou, G.; Poulou, M.; Malizos, K.; Stefanou, N.; Anastasopoulou, L.; Tsezou, A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin. Epigenetics 2017, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Yoshitomi, H.; Tanida, S.; Ishikawa, M.; Nishitani, K.; Ito, H.; Nakamura, T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 2010, 12, R86. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.; Gao, J.; Si, Y.; Zhao, D. Combination of circulating miR-19b-3p, miR-122-5p and miR-486-5p expressions correlates with risk and disease severity of knee osteoarthritis. Am. J. Transl. Res. 2017, 9, 2852–2864. [Google Scholar] [PubMed]
- Xie, W.; Su, W.; Xia, H.; Wang, Z.; Su, C.; Su, B. Synovial Fluid MicroRNA-210 as a Potential Biomarker for Early Prediction of Osteoarthritis. BioMed Res. Int. 2019, 2019, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Löfgren, S.E.; Frostegård, J.; Truedsson, L.; Pons-Estel, B.A.; D’Alfonso, S.; Witte, T.; Lauwerys, B.R.; Endreffy, E.; Kovács, L.; Vasconcelos, C.; et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012, 13, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Yang, W.; Ye, D.Q.; Cui, H.; Zhang, Y.; Hirankarn, N.; Qian, X.; Tang, Y.; Lau, Y.L.; de Vries, N.; et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011, 7, e1002128. [Google Scholar] [CrossRef]
- Sun, H.Y.; Lv, A.K.; Yao, H. Relationship of miRNA-146a to primary Sjögren’s syndrome and to systemic lupus erythematosus: A meta-analysis. Rheumatol. Int. 2017, 37, 1311–1316. [Google Scholar] [CrossRef]
- Li, J.; Yang, H.; Li, Y.; Liu, Y.; Chen, S.; Qi, C.; Zhang, Q.; Lan, T.; He, X.; Guan, X.Y.; et al. microRNA-146 up-regulation predicts the prognosis of non-small cell lung cancer by miRNA in situ hybridization. Exp. Mol. Pathol. 2014, 96, 195–199. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Sun, X.X.; Ma, X.; Chen, Z.N. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol. Cancer 2015, 14, 5. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, J.; Chu, J.; Yang, C.; Xiao, H.; Zhao, C.; Sun, Z.; Gao, X.; Chen, G.; Han, Z.; et al. MicroRNA-146a Induced by Hypoxia Promotes Chondrocyte Autophagy through Bcl-2. Cell. Physiol. Biochem. 2015, 37, 1442–1453. [Google Scholar] [CrossRef]
- Chen, G.; Gao, X.; Wang, J.; Yang, C.; Wang, Y.; Liu, Y.; Zou, W.; Liu, T. Hypoxia-induced microRNA-146a represses Bcl-2 through Traf6/IRAK1 but not Smad4 to promote chondrocyte autophagy. Biol. Chem. 2017, 398, 499–507. [Google Scholar] [CrossRef]
- Jin, L.; Zhao, J.; Jing, W.; Yan, S.; Wang, X.; Xiao, C.; Ma, B. Role of miR-146a in human chondrocyte apoptosis in response to mechanical pressure injury in vitro. Int. J. Mol. Med. 2014, 34, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Houard, X.; Goldring, M.B.; Berenbaum, F. Homeostatic Mechanisms in Articular Cartilage and Role of Inflammation in Osteoarthritis. Curr. Rheumatol. Rep. 2013, 15, 375. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Zhao, J.; Xu, J.; Geng, Y.; Dai, L.; Huang, Y.; Fu, S.C.; Dai, K.; Zhang, X. MiR-146a facilitates osteoarthritis by regulating cartilage homeostasis via targeting Camk2d and Ppp3r2. Cell Death Dis. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Tian, X.Y.; Huang, X.X.; He, L.L.; Xu, F. microRNA-186 inhibition of PI3K–AKT pathway via SPP1 inhibits chondrocyte apoptosis in mice with osteoarthritis. J. Cell. Physiol. 2019, 234, 6042–6053. [Google Scholar] [CrossRef] [PubMed]
- Miyaki, S.; Nakasa, T.; Otsuki, S.; Grogan, S.P.; Higashiyama, R.; Inoue, A.; Kato, Y.; Sato, T.; Lotz, M.K.; Asahara, H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009, 60, 2723–2730. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Zhuang, H.; Wang, G.; Li, Z.; Zhang, H.; Yu, T.; Zhang, B. MiRNA-140 is a negative feedback regulator of MMP-13 in IL-1β-stimulated human articular chondrocyte C28/I2 cells. Inflamm. Res. 2012, 61, 503–509. [Google Scholar] [CrossRef]
- Karlsen, T.A.; Jakobsen, R.B.; Mikkelsen, T.S.; Brinchmann, J.E. microRNA-140 Targets RALA and Regulates Chondrogenic Differentiation of Human Mesenchymal Stem Cells by Translational Enhancement of SOX9 and ACAN. Stem Cells Dev. 2014, 23, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Sondag, G.R.; Haqqi, T.M. The Role of MicroRNAs and Their Targets in Osteoarthritis. Curr. Rheumatol. Rep. 2016, 18, 56. [Google Scholar] [CrossRef]
- Patra, D.; Sandell, L.J. Recent advances in biomarkers in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 465–470. [Google Scholar] [CrossRef]
- Kurowska-Stolarska, M.; Alivernini, S.; Ballantine, L.E.; Asquith, D.L.; Millar, N.L.; Gilchrist, D.S.; Reilly, J.; Ierna, M.; Fraser, A.R.; Stolarski, B.; et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA 2011, 108, 11193–11198. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-F.; Zhou, Z.-H.; Zou, J. MicroRNA-181 inhibits proliferation and promotes apoptosis of chondrocytes in osteoarthritis by targeting PTEN. Biochem. Cell Biol. 2017, 95, 437–444. [Google Scholar] [CrossRef]
- Li, L.; Jia, J.; Liu, X.; Yang, S.; Ye, S.; Yang, W.; Zhang, Y. MicroRNA-16-5p Controls Development of Osteoarthritis by Targeting SMAD3 in Chondrocytes. Curr. Pharm. Des. 2015, 21, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, C.; Liu, X.; Yang, S.; Ye, S.; Jia, J.; Liu, W.; Zhang, Y. Elevated expression of microRNA-30b in osteoarthritis and its role in ERG regulation of chondrocyte. Biomed. Pharmacother. 2015, 76, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Song, H.; Wang, W.; Wang, H.; Peng, H.; Cui, J.; Wang, R.; Huang, H.; Wang, W.; Wang, L. Beclin 1 overexpression inhibits chondrocyte apoptosis and downregulates extracellular matrix metabolism in osteoarthritis. Mol. Med. Rep. 2017, 16, 3958–3964. [Google Scholar] [CrossRef]
- Yu, C.D.; Miao, W.H.; Zhang, Y.Y.; Zou, M.J.; Yan, X.F. Inhibition of miR-126 protects chondrocytes from IL-1β induced inflammation via upregulation of Bcl-2. Bone Jt. Res. 2018, 7, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Long, J.; Guo, W.; Ye, W. MicroRNA-195-5p inhibitor prevents the development of osteoarthritis by targeting REGγ. Mol. Med. Rep. 2019, 19, 4561–4568. [Google Scholar] [CrossRef]
- Prasadam, I.; Xiao, Y. A microRNA screen reveals the critical role of microRNA-23a-3p in maintaining cartilage homeostasis. Osteoarthr. Cartil. 2018, 26, S160. [Google Scholar] [CrossRef]
- Kang, L.; Yang, C.; Song, Y.; Liu, W.; Wang, K.; Li, S.; Zhang, Y. MicroRNA-23a-3p promotes the development of osteoarthritis by directly targeting SMAD3 in chondrocytes. Biochem. Biophys. Res. Commun. 2016, 478, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Philipot, D.; Guérit, D.; Platano, D.; Chuchana, P.; Olivotto, E.; Espinoza, F.; Dorandeu, A.; Pers, Y.M.; Piette, J.; Borzi, R.M.; et al. p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res. Ther. 2014, 16, R58. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Xu, J.; Chen, L.; Wu, H.; Feng, W.; Zheng, Y.; Li, P.; Zhang, H.; Zhang, L.; Chi, G.; et al. MicroRNA-27b targets CBFB to inhibit differentiation of human bone marrow mesenchymal stem cells into hypertrophic chondrocytes. Stem Cell Res. Ther. 2020, 11, 392. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Rasheed, Z.; Ramamurthy, S.; Anbazhagan, A.N.; Voss, F.R.; Haqqi, T.M. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010, 62, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Abouheif, M.M.; Nakasa, T.; Shibuya, H.; Niimoto, T.; Kongcharoensombat, W.; Ochi, M. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology 2010, 49, 2054–2060. [Google Scholar] [CrossRef]
- Luo, C.; Liang, J.S.; Gong, J.; Zhang, H.L.; Feng, Z.J.; Yang, H.T.; Zhang, H.B.; Kong, Q.H. The function of microRNA-34a in osteoarthritis. Bratisl. Med. J. 2019, 120, 386–391. [Google Scholar] [CrossRef]
- Mao, G.; Zhang, Z.; Huang, Z.; Chen, W.; Huang, G.; Meng, F.; Zhang, Z.; Kang, Y. MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr. Cartil. 2017, 25, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Xiaoling, G.; Shuaibin, L.; Kailu, L. MicroRNA-19b-3p promotes cell proliferation and osteogenic differentiation of BMSCs by interacting with lncRNA H19. BMC Med. Genet. 2020, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, F.; Song, Y.; Guan, X. miR-17-5p and miR-19b-3p prevent osteoarthritis progression by targeting EZH2. Exp. Ther. Med. 2020, 20, 1653–1663. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Guo, K.; Su, S.; Li, J.; Li, C. miR-486-5p is upregulated in osteoarthritis and inhibits chondrocyte proliferation and migration by suppressing SMAD2. Mol. Med. Rep. 2018, 18, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Kim, S.-J. Overexpression of MicroRNA-25 by Withaferin A Induces Cyclooxygenase-2 Expression in Rabbit Articular Chondrocytes. J. Pharmacol. Sci. 2014, 125, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.W.; Watkins, G.; Le Good, N.; Roberts, S.; Murphy, C.L.; Brockbank, S.M.V.; Needham, M.R.C.; Read, S.J.; Newham, P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-α and MMP13. Osteoarthr. Cartil. 2009, 17, 464–472. [Google Scholar] [CrossRef]
- Zhao, X.; Li, H.; Wang, L. MicroRNA-107 regulates autophagy and apoptosis of osteoarthritis chondrocytes by targeting TRAF3. Int. Immunopharmacol. 2019, 71, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Walayat, A.; Yang, M.; Xiao, D. Therapeutic Implication of miRNA in Human Disease. In Antisense Therapy; IntechOpen: London, UK, 2019. [Google Scholar]
- Oliviero, A.; Della Porta, G.; Peretti, G.M.; Maffulli, N. MicroRNA in osteoarthritis: Physiopathology, diagnosis and therapeutic challenge. Br. Med. Bull. 2019, 130, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Endisha, H.; Datta, P.; Sharma, A.; Nakamura, S.; Rossomacha, E.; Younan, C.; Ali, S.A.; Tavallaee, G.; Lively, S.; Potla, P.; et al. MicroRNA-34a-5p Promotes Joint Destruction during Osteoarthritis. Arthritis Rheumatol. 2020, 41552. [Google Scholar] [CrossRef]
- Yan, S.; Wang, M.; Zhao, J.; Zhang, H.; Zhou, C.; Jin, L.; Zhang, Y.; Qiu, X.; Ma, B.; Fan, Q. MicroRNA-34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int. J. Mol. Med. 2016, 38, 201–209. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, K.; Zhan, J.; Wu, M. miR-122/SIRT1 axis regulates chondrocyte extracellular matrix degradation in osteoarthritis. Biosci. Rep. 2020, 40, 40. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cai, L.; Zhang, Y.; Cui, L.; Shen, G. Intra-articular resveratrol injection prevents osteoarthritis progression in a mouse model by activating SIRT1 and thereby silencing HIF-2α. J. Orthop. Res. 2015, 33, 1061–1070. [Google Scholar] [CrossRef]
| Endogenous Factors | Comments |
| Age | Increasing incidence rate in younger adults. Over half of people diagnosed are under 65 years of age [10,11]. |
| Gender | OA is more common in women than men, particularly after menopausal age [12,13]. |
| Ethnicity | OA is more common in African-Americans compared to other ethnic groups [14]. |
| Genetics, Epigenetics | Genetic loci, multigene interaction, family heredity, methylation, histone modification, microRNA [7,8,15,16,17,18]. |
| Exogenous Factors | Comments |
| Knee events | Injury, joint surgery, knee pain, trauma or repeated loading [7,8,19]. |
| Obesity | BMI > 30 kg/m2 were 6.8 times more likely to develop knee OA than normal-weight controls [20]. |
| Lifestyle factors | Tobacco, alcohol assumption [21,22]. |
| Nutrition | Metabolic disease, such as lipid and cholesterol profiles, adequate vitamin levels, etc., essentially contribute to OA [23,24]. |
| Occupation, sports | The link between occupational activities, including kneeling, squatting, lifting, climbing, heavy stand working, heavy physical load work or combinations thereof, and OA has been reported [25,26,27]. Thus, many occupations have been reported to be linked to OA, such as floor layers, miners, dockers, carpenters, firefighting, mining asphalt, plumbers, bricklayers, sports at elite levels, etc. [28,29]. |
| References | c-miRNAs | Sources |
|---|---|---|
| [1] | miR-122↑, miR-25↓, miR-28-3p↓, miR-93↓, miR-140↓, miR-191↓, miR-342-3p↓, miR-146b↓, miR-454↓, miR-885-5p↑, miR-let-7b↓, miR-let-7e↓ | Serum |
| [50] | miR-146a↑, miR-155↑ | Peripheral blood |
| [51] | miR-132↓, miR-146a↑, miR-155↑, miR-181↑, miR-223↑ | Serum |
| [52] | miR-146a-5p↑ | Serum |
| [53] | miR-146a-5p↑, miR-186-5p↑ | Serum |
| [54] | miR-16↑, miR-20b↑, miR-19c↑, miR-30b↑, miR-93↑, miR-126↑, miR-184↑, miR-186↑, miR-195↑, miR-345↑, miR-885-5p↑ | Plasma |
| [55] * | miR-23a-3p↑, miR-24-3p↑, miR-27a-5p↓, miR-27b-3p↑, miR-29c-3p↑, miR-34a-5p↑, miR-329↓, miR-655↓, miR-708-3p↓, miR-934↓ and miR-186-5p↑ miR-27a-3p, miR-101-5p, miR-378-5p only detected in the late stage of OA. | Synovium |
| [56] | miR-140-3p↓, miR-33b-3p↓, miR-671-3p↓ | Serum |
| [57] | miR-132↓ | Synovium, plasma |
| [58] | miR-122-5p↑, miR-92a-3p↑, miR-19b-3p↑, miR-486-5p↑, miR-877-5p↓, miR-1180-3p↓, miR-320b↓, miR-663a↓ | Blood |
| [59] | miR-120↑ | Synovium |
| References | c-miRNAs | Target Genes | Function |
|---|---|---|---|
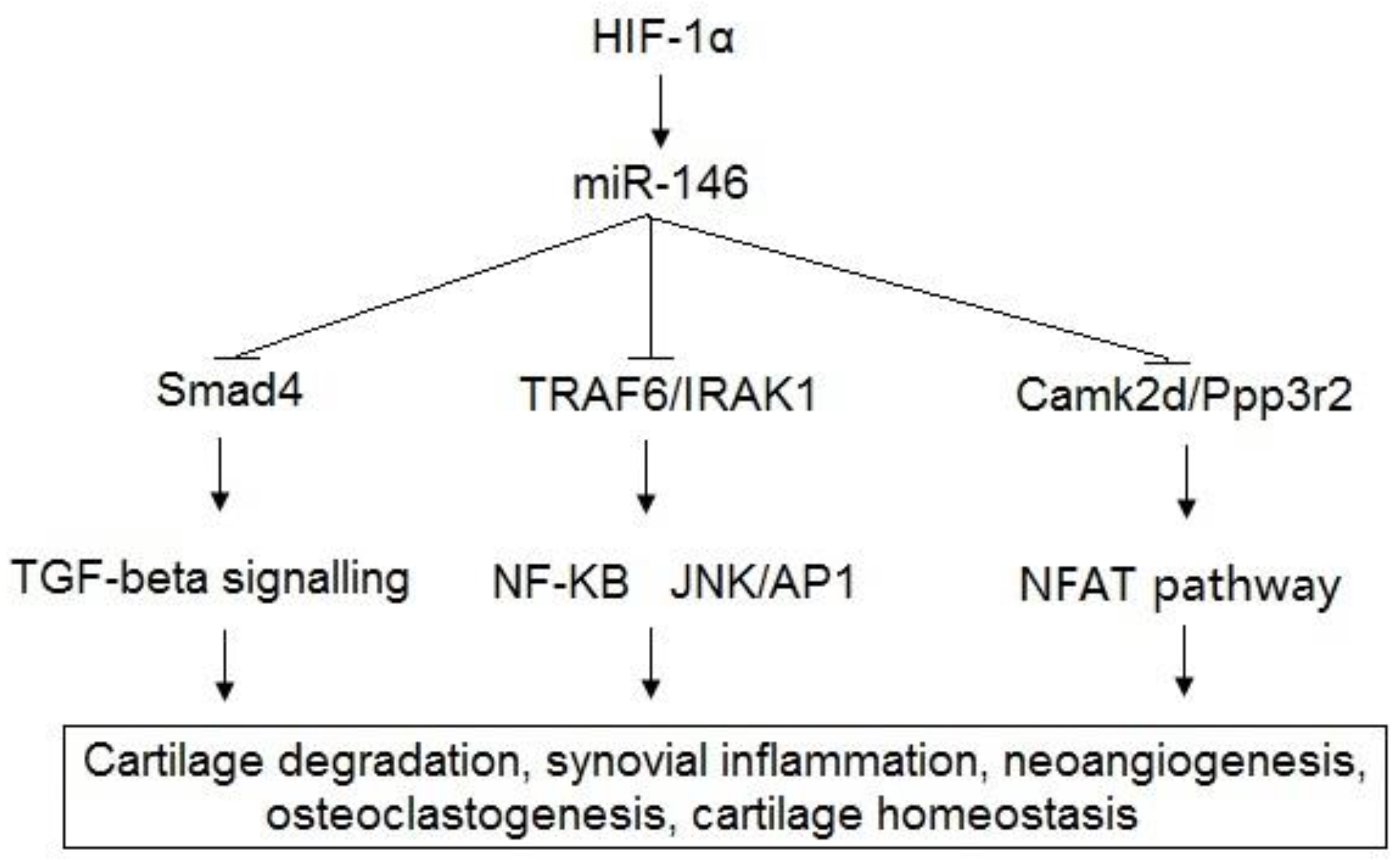
| [50,51,52,53,65,66,67,68,69] | miR-146a | Bcl-2, TRAF6, IRAK1, VEGF, Smad4, TGF-β, Camk2d, Ppp3r2 | Cartilage degradation, synovial inflammation, neoangiogenesis, osteoclastogenesis, cartilage homeostasis. |
| [37,41,76] | miR-155 | ULK1, MAP1LC3, ATG14, SHIP1 | Autophagy, inflammation |
| [77] | miR-181 | PTEN | Apoptosis |
| [78] | miR-16 | Smad3 | Chondrocyte growth, differentiation |
| [79,80] | miR-30b | ERG, BECN1 | Chondrocyte differentiation, autophagy |
| [81] | miR-126 | Bcl-2 | Inflammation |
| [70] | miR-186 | SPP1 | Chondrocyte apoptosis |
| [64,82] | miR-195 | HIF-1α, REGγ | Chondrocyte apoptosis, inflammation |
| [83,84] | miR-23a | Smad3, RUNX2 | Chondrocyte growth, cartilage homeostasis |
| [85] | miR-24 | p16INK4α | Reduces production of the two matrix remodeling enzymes |
| [86,87] | miR-27b | MMP-13, CBFB | Matrix degradation, chondrocyte differentiation |
| [88,89] | miR-34a | Col2α1, iNOS, TGIF2 | Chondrocyte apoptosis |
| [90] | miR-92a | HADC2 | Cartilage development and homeostasis |
| [91,92] | miR-19b | EZH2, LncRNA H19 | Chondrocyte apoptosis, ECM degradation |
| [93] | miR-486 | Smad2 | Chondrocyte growth |
| [56,71,72,73] | miR-140 | InsR, IGFR, ADAMTS5, MMP-13, IGFBP5, RALA | Metabolic processes, cartilage homeostasis and chondrogenesis. |
| [94,95] | miR-25 | COX2 | Inflammation |
| [96] | miR-107 | TRAF3 | Chondrocyte apoptosis, autophagy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lao, T.D.; Le, T.A.H. Data Integration Reveals the Potential Biomarkers of Circulating MicroRNAs in Osteoarthritis. Diagnostics 2021, 11, 412. https://doi.org/10.3390/diagnostics11030412
Lao TD, Le TAH. Data Integration Reveals the Potential Biomarkers of Circulating MicroRNAs in Osteoarthritis. Diagnostics. 2021; 11(3):412. https://doi.org/10.3390/diagnostics11030412
Chicago/Turabian StyleLao, Thuan Duc, and Thuy Ai Huyen Le. 2021. "Data Integration Reveals the Potential Biomarkers of Circulating MicroRNAs in Osteoarthritis" Diagnostics 11, no. 3: 412. https://doi.org/10.3390/diagnostics11030412
APA StyleLao, T. D., & Le, T. A. H. (2021). Data Integration Reveals the Potential Biomarkers of Circulating MicroRNAs in Osteoarthritis. Diagnostics, 11(3), 412. https://doi.org/10.3390/diagnostics11030412






