Comparable Accuracies of Nonstructural Protein 1- and Envelope Protein-Based Enzyme-Linked Immunosorbent Assays in Detecting Anti-Dengue Immunoglobulin G Antibodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Antibodies
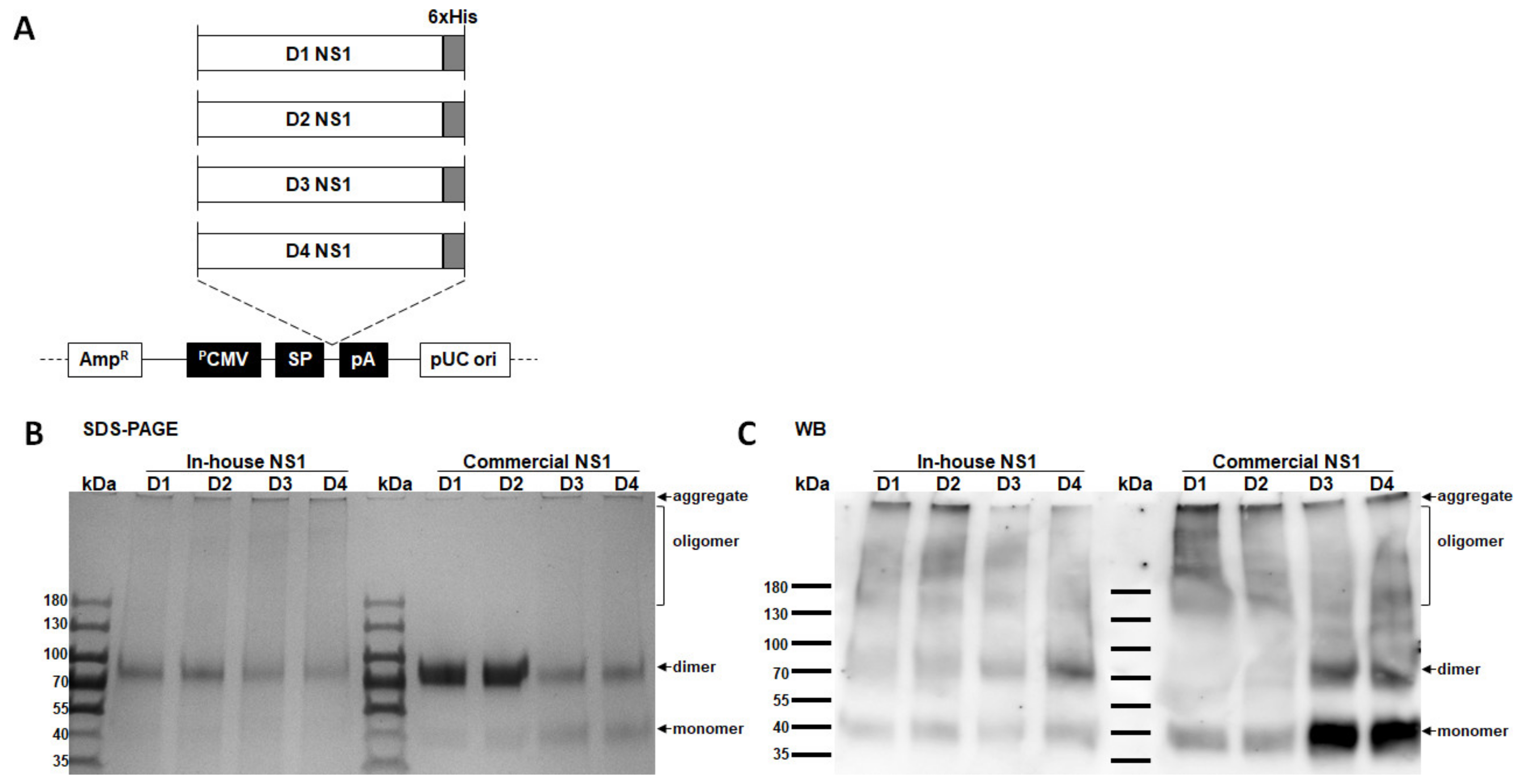
2.2. Plasmid Construction, Protein Expression, and Purification
2.3. Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS–PAGE) and Western Blotting
2.4. Generation of Stable Cell Lines
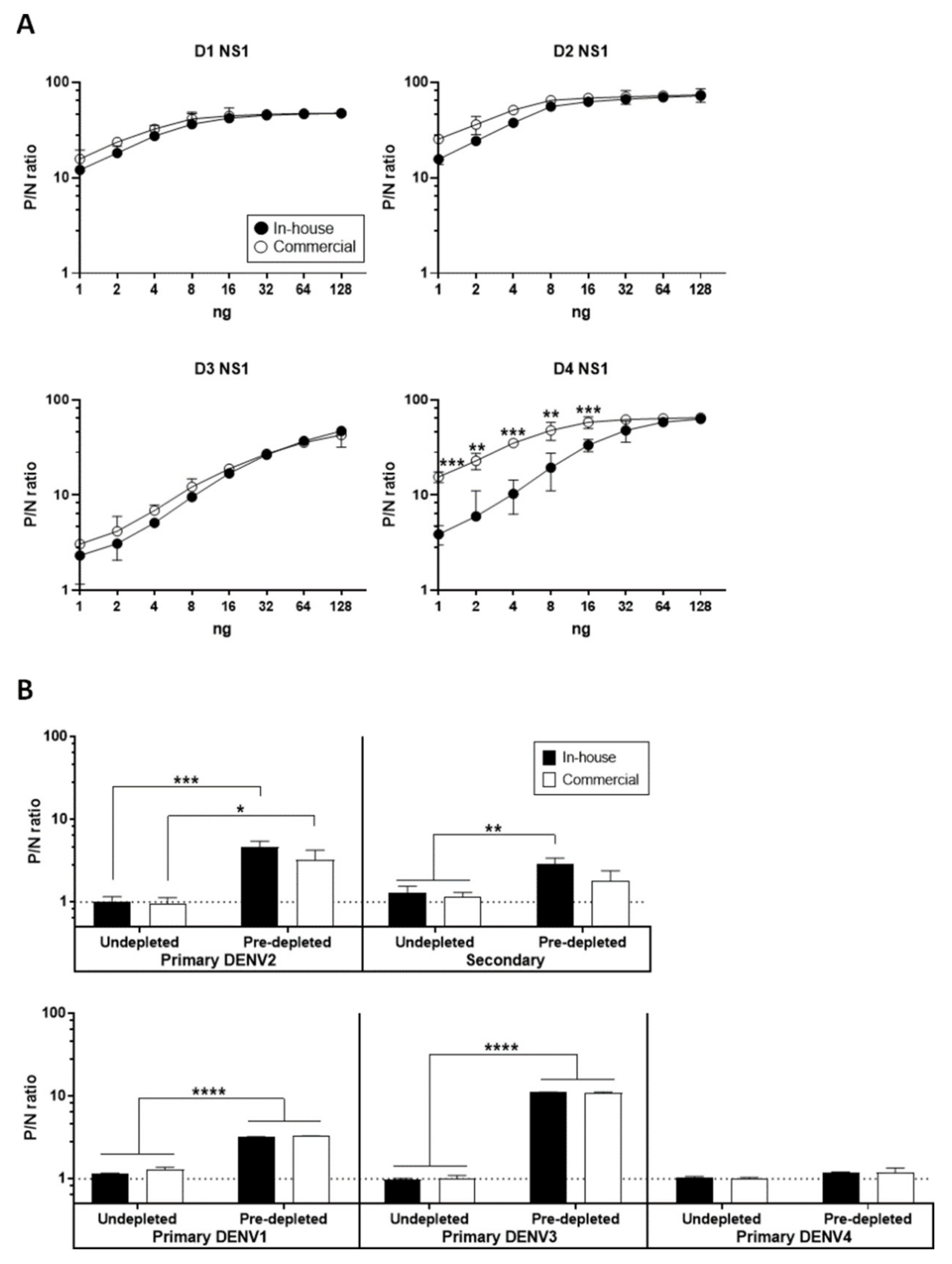
2.5. Antigen Titration by Ag–ELISA
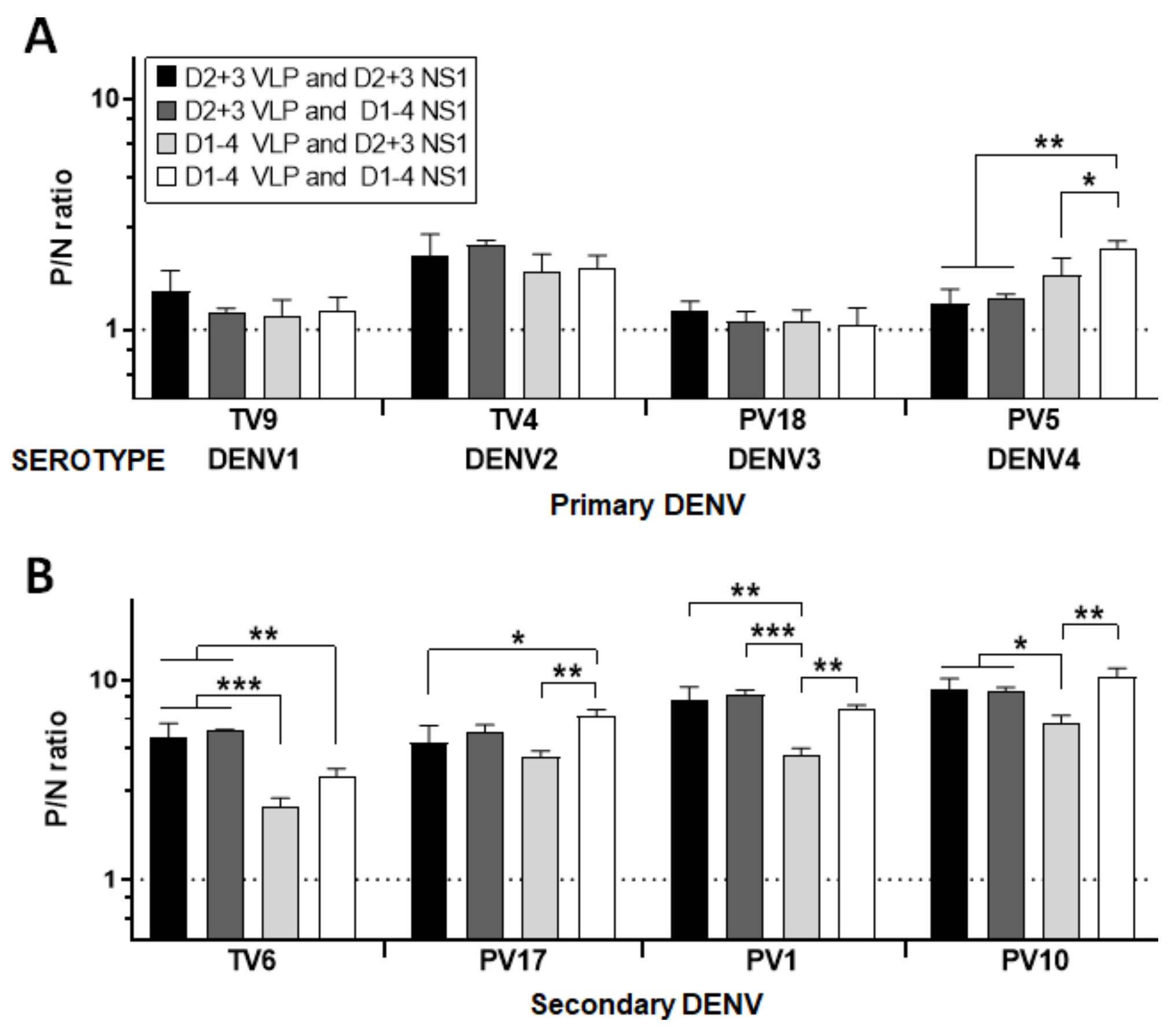
2.6. NS1- and VLP-Specific MAC/GAC–ELISAs
2.7. Indirect NS1 IgM/IgG ELISA
2.8. Limits of Detection of NS1 GAC- and Indirect NS1 IgG ELISAs
2.9. Human Serum Panel
2.10. Data Processing and Statistical Analysis
3. Results
3.1. Confirmation of In-House NS1 Protein Quality
3.2. Optimal Condition for NS1 GAC–ELISA
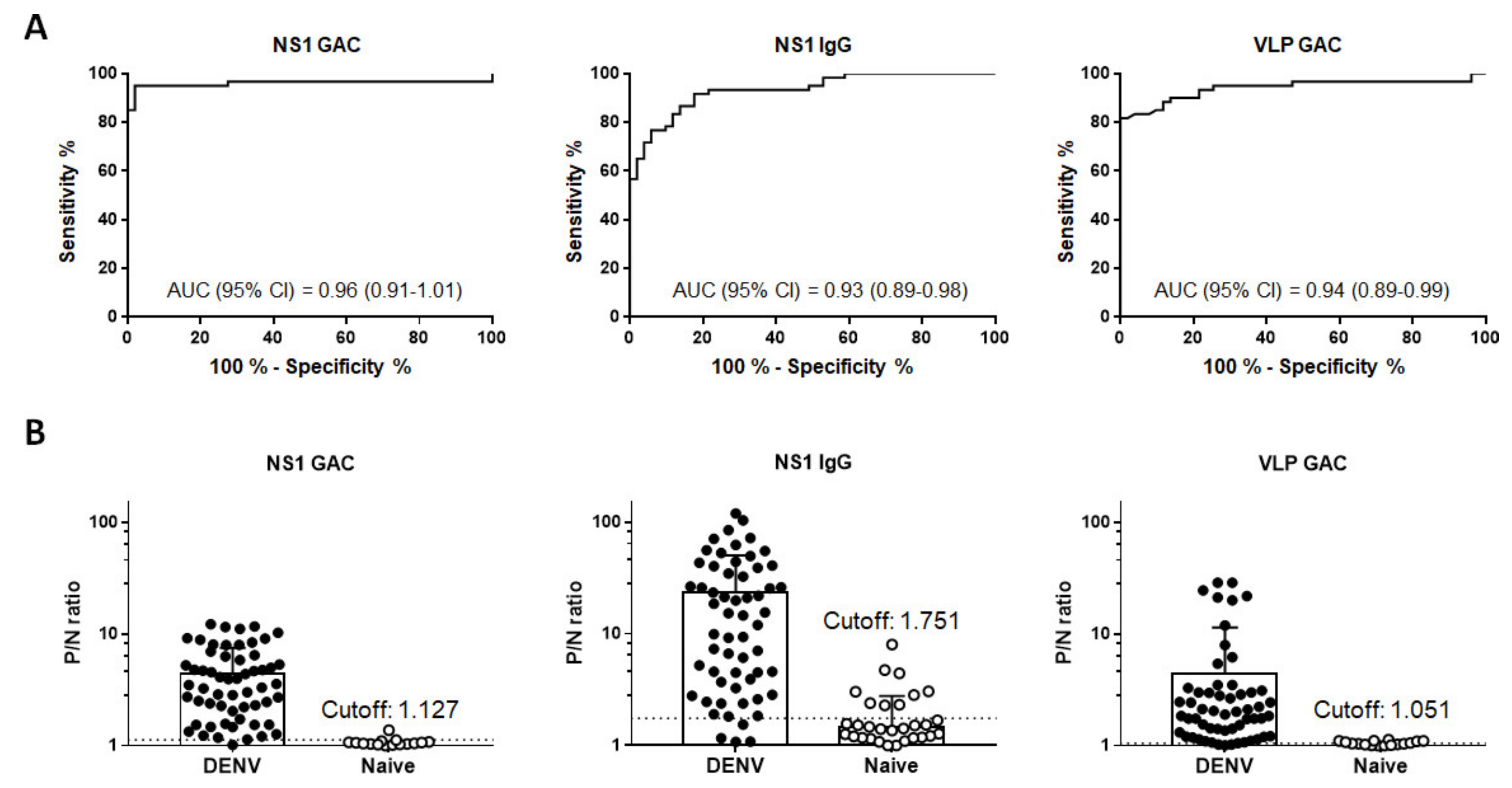
3.3. Overall Performance of Diagnostic Assays
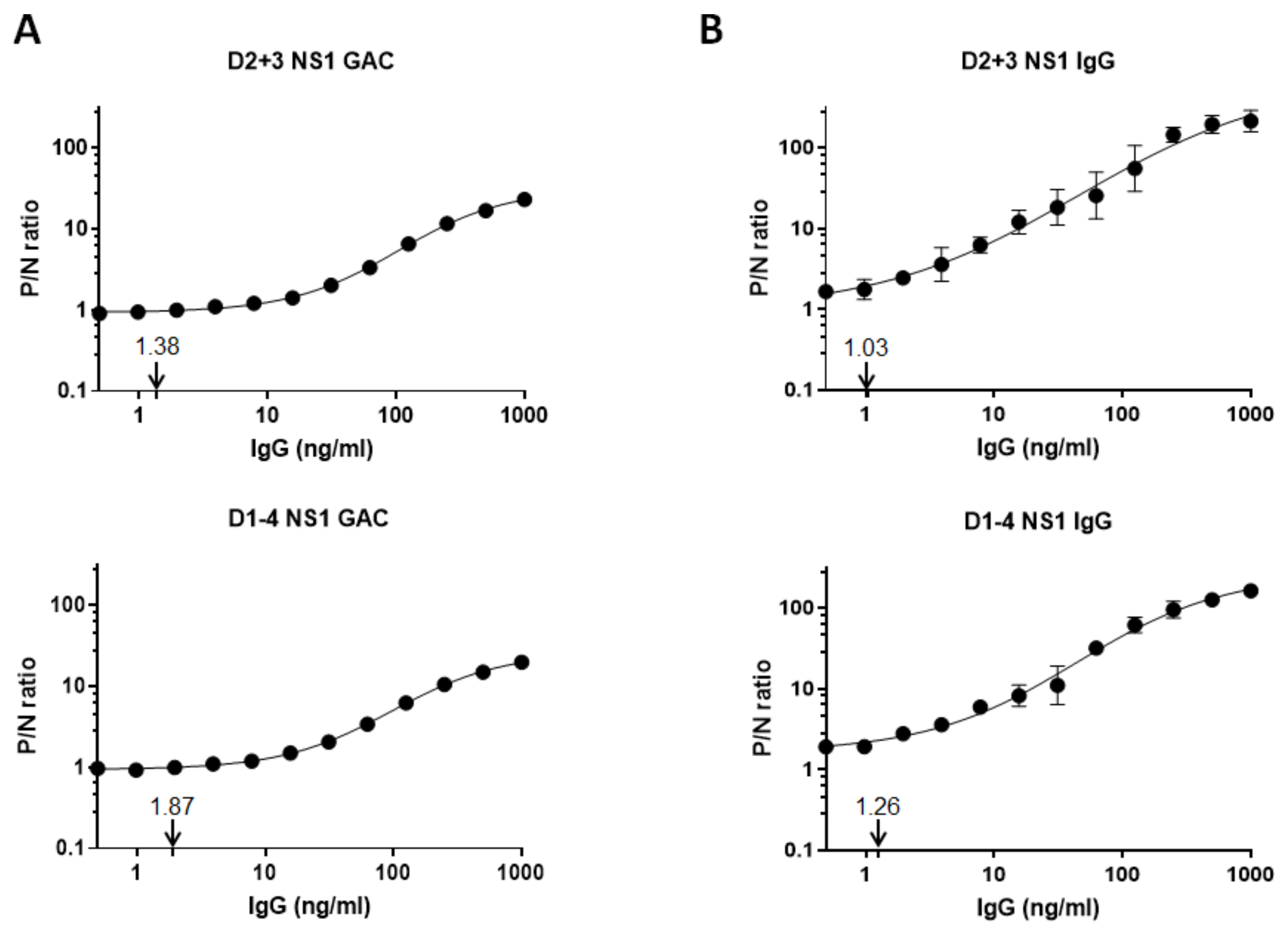
3.4. Limits of Anti-NS1 IgG Detection
3.5. Determination of Serostatus Using a Composite Reference Standard
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindenbach, B.D.M.; Murray, C.L.; Thiel, H.; Rice, C.M. Flaviviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 712–746. [Google Scholar]
- Calisher, C.H.; Karabatsos, N.; Dalrymple, J.M.; Shope, R.E.; Porterfield, J.S.; Westaway, E.G.; Brandt, W.E. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 1989, 70 Pt 1, 37–43. [Google Scholar] [CrossRef]
- Halstead, S.B. Dengue. Lancet 2007, 370, 1644–1652. [Google Scholar] [CrossRef]
- Kyle, J.L.; Harris, E. Global spread and persistence of dengue. Annu. Rev. Microbiol. 2008, 62, 71–92. [Google Scholar] [CrossRef] [Green Version]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martinez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Ishikawa, T.; Yamanaka, A.; Konishi, E. A review of successful flavivirus vaccines and the problems with those flaviviruses for which vaccines are not yet available. Vaccine 2014, 32, 1326–1337. [Google Scholar] [CrossRef]
- Collins, M.H.; Metz, S.W. Progress and works in progress: Update on flavivirus vaccine development. Clin. Ther. 2017, 39, 1519–1536. [Google Scholar] [CrossRef] [Green Version]
- Galula, J.U.; Salem, G.M.; Chang, G.J.; Chao, D.Y. Does structurally-mature dengue virion matter in vaccine preparation in post-Dengvaxia era? Hum. Vaccin. Immunother. 2019, 15, 2328–2336. [Google Scholar] [CrossRef] [Green Version]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [Green Version]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003, 59, 23–61. [Google Scholar] [CrossRef]
- Schlesinger, J.J.; Brandriss, M.W.; Walsh, E.E. Protection of mice against dengue 2 virus encephalitis by immunization with the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 1987, 68 Pt 3, 853–857. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henchal, E.A.; Henchal, L.S.; Schlesinger, J.J. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J. Gen. Virol. 1988, 69 Pt 8, 2101–2107. [Google Scholar] [CrossRef]
- Albinsson, B.; Vene, S.; Rombo, L.; Blomberg, J.; Lundkvist, A.; Ronnberg, B. Distinction between serological responses following tick-borne encephalitis virus (TBEV) infection vs vaccination, Sweden 2017. Euro Surveill. 2018, 23. [Google Scholar] [CrossRef]
- Konishi, E.; Suzuki, T. Ratios of subclinical to clinical Japanese encephalitis (JE) virus infections in vaccinated populations: Evaluation of an inactivated JE vaccine by comparing the ratios with those in unvaccinated populations. Vaccine 2002, 21, 98–107. [Google Scholar] [CrossRef]
- Konishi, E. Status of natural infection with Japanese encephalitis virus in Japan: Prevalence of antibodies to the nonstructural 1 protein among humans and horses. Vaccine 2009, 27, 7129–7130. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.Y.; Chen, L.K.; Chang, S.F.; Yueh, Y.Y.; Chow, L.; Chien, L.J.; Chin, C.; Lin, T.H.; Huang, J.H. Antibody to the nonstructural protein NS1 of Japanese encephalitis virus: Potential application of mAb-based indirect ELISA to differentiate infection from vaccination. Vaccine 2001, 19, 1753–1763. [Google Scholar] [CrossRef]
- Rothman, A.L. Immunity to dengue virus: A tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 2011, 11, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, E.J.M.; George, J.K.; Velasco, M.; Bonaparte, M.I.; Zheng, L.; DiazGranados, C.A.; Marques, E.T.A.; Huleatt, J.W. Development of an anti-dengue NS1 IgG ELISA to evaluate exposure to dengue virus. J. Virol. Methods 2018, 257, 48–57. [Google Scholar] [CrossRef]
- Nascimento, E.J.M.; Huleatt, J.W.; Cordeiro, M.T.; Castanha, P.M.S.; George, J.K.; Grebe, E.; Welte, A.; Brown, M.; Burke, D.S.; Marques, E.T.A. Development of antibody biomarkers of long term and recent dengue virus infections. J. Virol. Methods 2018, 257, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.Y.; Chen, L.K.; Chang, S.F.; Yueh, Y.Y.; Chow, L.; Chien, L.J.; Chin, C.; Yang, H.H.; Lin, T.H.; Huang, J.H. Potential application of nonstructural protein NS1 serotype-specific immunoglobulin G enzyme-linked immunosorbent assay in the seroepidemiologic study of dengue virus infection: Correlation of results with those of the plaque reduction neutralization test. J. Clin. Microbiol. 2002, 40, 1840–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyson, J.; Tsai, W.Y.; Tsai, J.J.; Brites, C.; Massgard, L.; Ha Youn, H.; Pedroso, C.; Drexler, J.F.; Stramer, S.L.; Balmaseda, A.; et al. Combination of nonstructural protein 1-based enzyme-linked immunosorbent assays can detect and distinguish various dengue virus and zika virus infections. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, P.Y.; Chen, L.K.; Chang, S.F.; Yueh, Y.Y.; Chow, L.; Chien, L.J.; Chin, C.; Lin, T.H.; Huang, J.H. Dengue NS1-specific antibody responses: Isotype distribution and serotyping in patients with dengue fever and dengue hemorrhagic fever. J. Med. Virol. 2000, 62, 224–232. [Google Scholar] [CrossRef]
- Konishi, E.; Shoda, M.; Ajiro, N.; Kondo, T. Development and evaluation of an enzyme-linked immunosorbent assay for quantifying antibodies to Japanese encephalitis virus nonstructural 1 protein to detect subclinical infections in vaccinated horses. J. Clin. Microbiol. 2004, 42, 5087–5093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, P.Y.; Chen, L.K.; Chang, S.F.; Su, C.L.; Chien, L.J.; Chin, C.; Lin, T.H.; Huang, J.H. Dengue virus serotyping based on envelope and membrane and nonstructural protein NS1 serotype-specific capture immunoglobulin M enzyme-linked immunosorbent assays. J. Clin. Microbiol. 2004, 42, 2489–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, D.Y.; Galula, J.U.; Shen, W.F.; Davis, B.S.; Chang, G.J. Nonstructural protein 1-specific immunoglobulin M and G antibody capture enzyme-linked immunosorbent assays in diagnosis of flaviviral infections in humans. J. Clin. Microbiol. 2015, 53, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Galula, J.U.; Chang, G.J.; Chuang, S.T.; Chao, D.Y. Establishment of an algorithm using prM/E- and NS1-specific IgM antibody-capture enzyme-linked immunosorbent assays in diagnosis of Japanese encephalitis virus and West Nile virus infections in humans. J. Clin. Microbiol. 2016, 54, 412–422. [Google Scholar] [CrossRef] [Green Version]
- Chao, D.Y.; Whitney, M.T.; Davis, B.S.; Medina, F.A.; Munoz, J.L.; Chang, G.J. Comprehensive evaluation of differential serodiagnosis between zika and dengue viral infections. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.J.; Hunt, A.R.; Holmes, D.A.; Springfield, T.; Chiueh, T.S.; Roehrig, J.T.; Gubler, D.J. Enhancing biosynthesis and secretion of premembrane and envelope proteins by the chimeric plasmid of dengue virus type 2 and Japanese encephalitis virus. Virology 2003, 306, 170–180. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.J.; Davis, B.S.; Stringfield, C.; Lutz, C. Prospective immunization of the endangered California condors (Gymnogyps californianus) protects this species from lethal West Nile virus infection. Vaccine 2007, 25, 2325–2330. [Google Scholar] [CrossRef]
- Holmes, D.A.; Purdy, D.E.; Chao, D.Y.; Noga, A.J.; Chang, G.J. Comparative analysis of immunoglobulin M (IgM) capture enzyme-linked immunosorbent assay using virus-like particles or virus-infected mouse brain antigens to detect IgM antibody in sera from patients with evident flaviviral infections. J. Clin. Microbiol. 2005, 43, 3227–3236. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Gammell, P.; Clynes, M. Proliferation control strategies to improve productivity and survival during CHO based production culture: A summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology 2007, 53, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Crill, W.D.; Hughes, H.R.; Delorey, M.J.; Chang, G.J. Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS ONE 2009, 4, e4991. [Google Scholar] [CrossRef]
- Shen, W.F.; Galula, J.U.; Liu, J.H.; Liao, M.Y.; Huang, C.H.; Wang, Y.C.; Wu, H.C.; Liang, J.J.; Lin, Y.L.; Whitney, M.T.; et al. Epitope resurfacing on dengue virus-like particle vaccine preparation to induce broad neutralizing antibody. Elife 2018, 7. [Google Scholar] [CrossRef]
- Quinn, C.P.; Semenova, V.A.; Elie, C.M.; Romero-Steiner, S.; Greene, C.; Li, H.; Stamey, K.; Steward-Clark, E.; Schmidt, D.S.; Mothershed, E.; et al. Specific, sensitive, and quantitative enzyme-linked immunosorbent assay for human immunoglobulin G antibodies to anthrax toxin protective antigen. Emerg. Infect. Dis. 2002, 8, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Makela, O.; Karjalainen, K. Inherited immunoglobulin idiotypes of the mouse. Immunol. Rev. 1977, 34, 119–138. [Google Scholar] [CrossRef]
- Benkirane, N.; Guichard, G.; Van Regenmortel, M.H.; Briand, J.P.; Muller, S. Cross-reactivity of antibodies to retro-inverso peptidomimetics with the parent protein histone H3 and chromatin core particle. Specificity and kinetic rate-constant measurements. J. Biol. Chem. 1995, 270, 11921–11926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puschnik, A.; Lau, L.; Cromwell, E.A.; Balmaseda, A.; Zompi, S.; Harris, E. Correlation between dengue-specific neutralizing antibodies and serum avidity in primary and secondary dengue virus 3 natural infections in humans. PLoS Negl. Trop. Dis. 2013, 7, e2274. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.Y.; Williams, K.L.; Wu, Y.C.; Knight, S.; Balmaseda, A.; Harris, E.; Wang, W.K. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in Nicaraguan dengue cases. PLoS Negl. Trop. Dis. 2013, 7, e2451. [Google Scholar] [CrossRef]
- Raafat, N.; Blacksell, S.D.; Maude, R.J. A review of dengue diagnostics and implications for surveillance and control. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 653–660. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Fongwen, N.; Kelly-Cirino, C.; Harris, E.; Wilder-Smith, A.; Peeling, R.W. Rapid diagnostic tests for determining dengue serostatus: A systematic review and key informant interviews. Clin. Microbiol. Infect. 2019, 25, 659–666. [Google Scholar] [CrossRef] [Green Version]
- Konishi, E.; Kitai, Y. Detection by ELISA of antibodies to Japanese encephalitis virus nonstructural 1 protein induced in subclinically infected humans. Vaccine 2009, 27, 7053–7058. [Google Scholar] [CrossRef]
- Tsai, W.Y.; Youn, H.H.; Brites, C.; Tsai, J.J.; Tyson, J.; Pedroso, C.; Drexler, J.F.; Stone, M.; Simmons, G.; Busch, M.P.; et al. Distinguishing secondary dengue virus infection from zika virus infection with previous dengue by a combination of 3 simple serological tests. Clin. Infect. Dis. 2017, 65, 1829–1836. [Google Scholar] [CrossRef]
- Wong, S.J.; Furuya, A.; Zou, J.; Xie, X.; Dupuis, A.P., II; Kramer, L.D.; Shi, P.Y. A multiplex microsphere immunoassay for zika virus diagnosis. EBioMedicine 2017, 16, 136–140. [Google Scholar] [CrossRef] [Green Version]
- De Alwis, R.; Beltramello, M.; Messer, W.B.; Sukupolvi-Petty, S.; Wahala, W.M.; Kraus, A.; Olivarez, N.P.; Pham, Q.; Brien, J.D.; Tsai, W.Y.; et al. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl. Trop. Dis. 2011, 5, e1188. [Google Scholar] [CrossRef]
- Beltramello, M.; Williams, K.L.; Simmons, C.P.; Macagno, A.; Simonelli, L.; Quyen, N.T.; Sukupolvi-Petty, S.; Navarro-Sanchez, E.; Young, P.R.; de Silva, A.M.; et al. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010, 8, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Guy, B.; Jackson, N. Dengue vaccine: Hypotheses to understand CYD-TDV-induced protection. Nat. Rev. Microbiol. 2016, 14, 45–54. [Google Scholar] [CrossRef]
- Hadinegoro, S.R.; Arredondo-Garcia, J.L.; Capeding, M.R.; Deseda, C.; Chotpitayasunondh, T.; Dietze, R.; Muhammad Ismail, H.I.; Reynales, H.; Limkittikul, K.; Rivera-Medina, D.M.; et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N. Engl. J. Med. 2015, 373, 1195–1206. [Google Scholar] [CrossRef] [Green Version]
- Capeding, M.R.; Tran, N.H.; Hadinegoro, S.R.; Ismail, H.I.; Chotpitayasunondh, T.; Chua, M.N.; Luong, C.Q.; Rusmil, K.; Wirawan, D.N.; Nallusamy, R.; et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: A phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 2014, 384, 1358–1365. [Google Scholar] [CrossRef]
- Rodriguez-Barraquer, I.; Mier, Y.T.-R.L.; Ferguson, N.; Burke, D.S.; Cummings, D.A.T. Differential efficacy of dengue vaccine by immune status. Lancet 2015, 385, 1726. [Google Scholar] [CrossRef]
- Sridhar, S.; Luedtke, A.; Langevin, E.; Zhu, M.; Bonaparte, M.; Machabert, T.; Savarino, S.; Zambrano, B.; Moureau, A.; Khromava, A.; et al. Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med. 2018, 379, 327–340. [Google Scholar] [CrossRef]
- World Health Organization. Dengue vaccine: WHO position paper, September 2018—Recommendations. Vaccine 2018, 37, 4848–4849. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Ooi, E.E.; Horstick, O.; Wills, B. Dengue. Lancet 2019, 393, 350–363. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Smith, P.G.; Luo, R.; Kelly-Cirino, C.; Curry, D.; Larson, H.; Durbin, A.; Chu, M.; Tharmaphornpilas, P.; Ng, L.C.; et al. Pre-vaccination screening strategies for the use of the CYD-TDV dengue vaccine: A meeting report. Vaccine 2019, 37, 5137–5146. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.L.; Adams, C.; Ylade, M.; Jadi, R.; Daag, J.V.; Molloy, C.T.; Agrupis, K.A.; Kim, D.R.; Silva, M.W.; Yoon, I.K.; et al. Determining dengue virus serostatus by indirect IgG ELISA compared with focus reduction neutralisation test in children in Cebu, Philippines: A prospective population-based study. Lancet Glob. Health 2021, 9, e44–e51. [Google Scholar] [CrossRef]
- Kosack, C.S.; Page, A.L.; Klatser, P.R. A guide to aid the selection of diagnostic tests. Bull. World Health Organ. 2017, 95, 639–645. [Google Scholar] [CrossRef]
- Land, K.J.; Boeras, D.I.; Chen, X.S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
| Type of Assay | Serum Panel | Classification | No. of Samples | Confirmatory Test |
|---|---|---|---|---|
| NS1 a and VLP b GAC c- and Indirect NS1 IgG ELISA | Community sera | |||
| Primary DENV1 | Late convalescent f | 7 | FRµNT90 * | |
| Primary DENV2 | Late convalescent | 10 | FRµNT90 | |
| Primary DENV3 | Late convalescent | 2 | FRµNT90 | |
| Primary DENV4 | Late convalescent | 1 | FRµNT90 | |
| Secondary infections | Late convalescent | 41 | FRµNT90 | |
| NS1 and VLP MAC d- and Indirect NS1 IgM ELISA e | Patient sera | |||
| Primary DENV1 | Acute g | 5 | RT–PCR # | |
| Early convalescent h | 7 | |||
| Late convalescent | 5 | |||
| Primary DENV2 | Early convalescent | 2 | RT–PCR | |
| Primary DENV3 | Acute | 1 | RT–PCR | |
| Early convalescent | 11 | |||
| NS1 and VLP GAC/MAC- and Indirect NS1 IgG/IgM ELISA | Naïve sera | n/a | 51 | FRµNT90 |
| FRµNT | ||||||
|---|---|---|---|---|---|---|
| Test | Result | Positive | Negative | AUC # (95% CI) | % Sensitivity (95% CI) | % Specificity (95% CI) |
| NS1 GAC | Positive | 57 | 1 | 0.96 (0.91–1.01) a,b | 95 (86.08–98.96) | 98.04 (89.55–99.95) |
| Negative | 3 | 50 | ||||
| NS1 IgG | Positive | 55 | 9 | 0.93 (0.89–0.98) c | 91.67 (81.61–97.24) | 82.35 (69.13–91.60) |
| Negative | 5 | 42 | ||||
| VLP GAC | Positive | 54 | 7 | 0.94 (0.89–0.99) | 90 (79.49–96.24) | 86.27 (73.74–94.30) |
| Negative | 6 | 44 | ||||
| Composite reference standard 1 * | Positive | 57 | 2 | n/a | 95 (86.08–98.96) | 96.08 (86.54–99.52) |
| Negative | 3 | 49 | ||||
| Composite reference standard 2 ** | Positive | 51 | 0 | n/a | 85 (73.43–92.90) | 100 (96.38–1.00) |
| Negative | 9 | 51 | ||||
| Reference | Type of Assay | Serum Dilution | Antigens Used | Gold Standard | % Sensitivity (95% CI) (or % Positive Rate) | % Specificity (95% CI) | MDC a |
|---|---|---|---|---|---|---|---|
| [21] | Indirect IgG ELISA | 1:50 | DENV1–4 NS1 | PRNT c | 91.89 (83.11–96.54) | 84.62 (65.85–94.47) | 2.33 EU/ml |
| [24] | Indirect IgG ELISA | 1:400 | DENV1–4 NS1 | RT–PCR d | 95.6 (91.40–97.80) | 89.5 (84.10–92.30) | n/a |
| [45] | Indirect IgG ELISA | 1:400 | DENV1 NS1 | RT–PCR | 81.30–95.80 positive rate | n/a e | n/a |
| [46] | MIA b | 1:100 | DENV1–4 NS1 | PRNT | 89 (54.33–99.99) | 86 (46.65–99.47) | n/a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galula, J.U.; Salem, G.M.; Destura, R.V.; Remenyi, R.; Chao, D.-Y. Comparable Accuracies of Nonstructural Protein 1- and Envelope Protein-Based Enzyme-Linked Immunosorbent Assays in Detecting Anti-Dengue Immunoglobulin G Antibodies. Diagnostics 2021, 11, 741. https://doi.org/10.3390/diagnostics11050741
Galula JU, Salem GM, Destura RV, Remenyi R, Chao D-Y. Comparable Accuracies of Nonstructural Protein 1- and Envelope Protein-Based Enzyme-Linked Immunosorbent Assays in Detecting Anti-Dengue Immunoglobulin G Antibodies. Diagnostics. 2021; 11(5):741. https://doi.org/10.3390/diagnostics11050741
Chicago/Turabian StyleGalula, Jedhan Ucat, Gielenny M. Salem, Raul V. Destura, Roland Remenyi, and Day-Yu Chao. 2021. "Comparable Accuracies of Nonstructural Protein 1- and Envelope Protein-Based Enzyme-Linked Immunosorbent Assays in Detecting Anti-Dengue Immunoglobulin G Antibodies" Diagnostics 11, no. 5: 741. https://doi.org/10.3390/diagnostics11050741






