Genetic Basis of Dual Diagnosis: A Review of Genome-Wide Association Studies (GWAS) Focusing on Patients with Mood or Anxiety Disorders and Co-Occurring Alcohol-Use Disorders
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Algorithm
2.1.1. Inclusion Criteria
- Articles written in English and published in peer-reviewed journals;
- Studies performed in humans (animal models relevant to human findings were allowed);
- Studies of samples with phenotypes of interest—MDD/AUD, BPD/AUD or Anxiety/Anxiety Disorder/AUD identifying the presence of SNPs with a genome-wide level of significance (p < 5 × 10−8) or suggestive genome-wide level of significance (p < 1 × 10−4);
- Papers reporting statistically significant correlation between MDD-PRS, BPD-PRS or Anxiety/Neuroticism PRS and alcohol phenotypes—DSM-IV alcohol dependence or alcohol abuse.
2.1.2. Exclusion Criteria
- Studies including alcohol phenotypes that are not based on DSM-IV/5 or ICD-10 criteria, but on screening or other tools for assessment of alcohol use instead—e.g., Alcohol Use Disorders Identification Test (AUDIT) [42].
2.2. Data Sources and Keywords
2.3. Selection of Studies
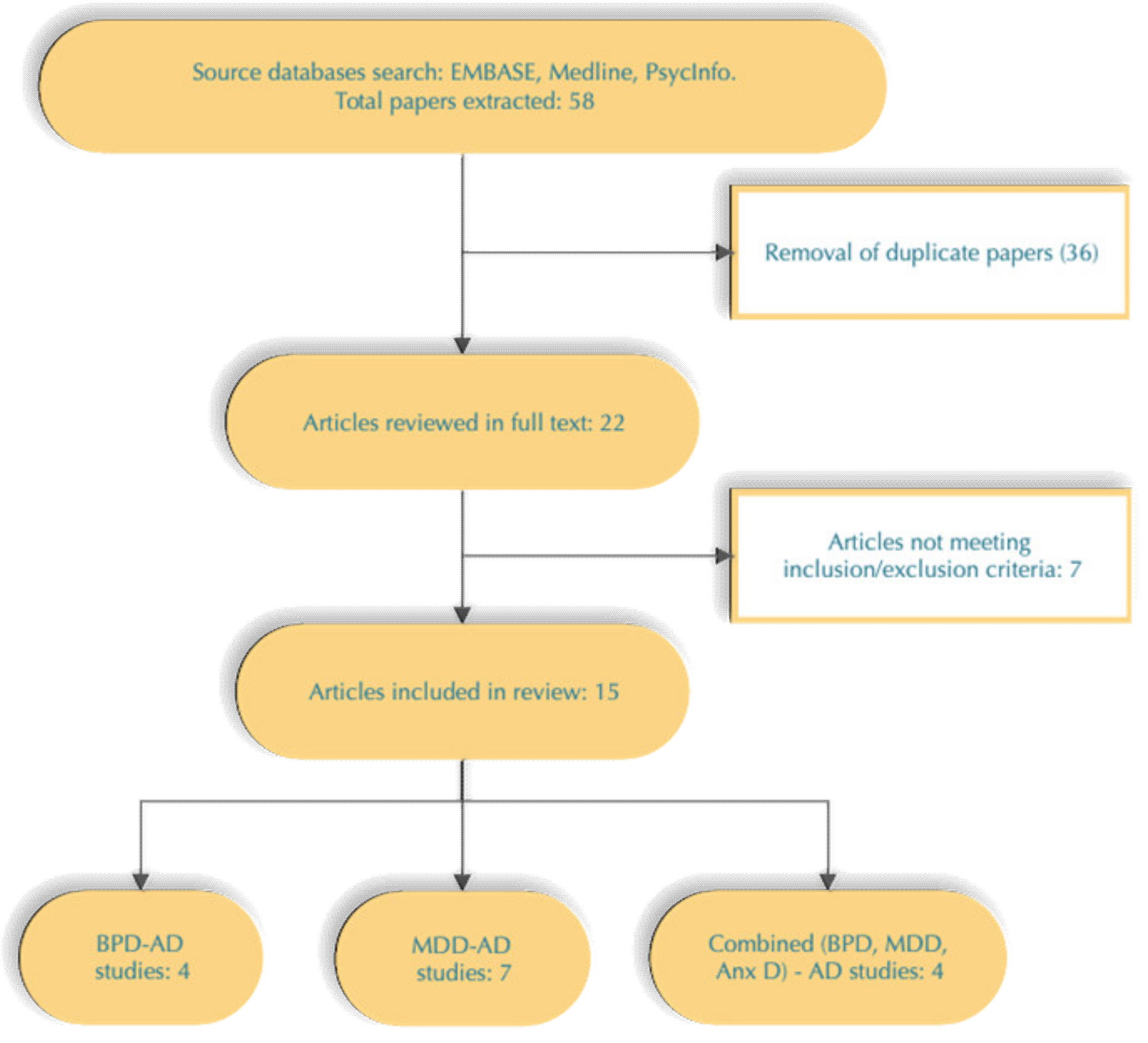
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Griswold, M.G.; Fullman, N.; Hawley, C.; Arian, N.; Zimsen, S.R.M.; Tymeson, H.D.; Venkateswaran, V.; Tapp, A.D.; Forouzanfar, M.H.; Salama, J.S.; et al. Alcohol use and burden for 195 countries and territories, 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet 2018, 392, 1015–1035. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Depression and Other Common Mental Disorders; WHO: Geneve, Switzerland, 2017. [Google Scholar]
- Grant, B.F.; Stinson, F.S.; Dawson, D.A.; Chou, P.; Dufour, M.C.; Compton, W.; Pickenberg, R.F.; Kaplan, K. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety disorders: Results from the national epidemiologic survey on alcohol and related conditions. Arch. Gen. Psychiatry 2004, 61, 807–816. [Google Scholar] [CrossRef]
- Boschloo, L.; Vogelzangs, N.; Van den Brink, W.; Smit, J.; Veltman, D.; Beekman, A.T.; Pennix, B.W. Alcohol use disorders and the course of depressive and anxiety disorders. Br. J. Psychiatry 2012, 200, 476–484. [Google Scholar] [CrossRef]
- Prior, K.; Mills, K.; Ross, J.; Teesson, M. Substance use disorders comorbid with mood and anxiety disorders in the Australian general population. Drug Alc. Rev. 2016, 36, 317–324. [Google Scholar] [CrossRef]
- Kessler, R.C.; Crum, R.M.; Warner, L.A.; Nelson, C.B.; Schulenberg, J.; Anthony, J.C. Lifetime co-occurrence of DSM-III-R alcohol abuse and dependence with other psychiatric disorders in the National Comorbidity Survey. Arch. Gen. Psychiatry 1997, 54, 313–321. [Google Scholar] [CrossRef]
- Offord, D.R.; Boyle, M.H.; Campbell, D.; Goering, P.; Lin, E.; Wong, M.; Racine, Y.A. One-year prevalence of psychiatric disorder in Ontarians 15 to 64 years of age. Can. J. Psychiatry 1996, 41, 559–563. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, R.; Bijl, R.V.; Smit, F.; Vollebergh, W.A.; Spijker, J. Risk factors for 12-month comorbidity of mood, anxiety, and substance use disorders: Findings from the Netherlands Mental Health Survey and Incidence Study. Am. J. Psychiatry 2002, 159, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Hasin, D.S.; Stinson, F.S.; Ogburn, E.; Grant, B.F. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch. Gen. Psychiatry 2007, 64, 830–842. [Google Scholar] [CrossRef]
- Conway, K.P.; Compton, W.; Stinson, F.S.; Grant, B.F. Lifetime comorbidity of DSM-IV mood and anxiety disorders and specific drug use disorders: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. J. Clin. Psychiatry 2006, 67, 247–257. [Google Scholar] [CrossRef]
- Kuria, M.W.; Ndetei, D.M.; Obot, I.S.; Khasakhala, L.I.; Bagaka, B.M.; Mbugua, M.N.; Kamau, J. The association between alcohol dependence and depression before and after treatment for alcohol dependence. ISRN Psychiatry 2012, 482802. [Google Scholar] [CrossRef]
- Gabriels, C.M.; Macharia, M.; Weich, L. Psychiatric comorbidity among alcohol-dependent individuals seeking treatment at the Alcohol Rehabilitation Unit, Stikland Hospital. S. Afr. J. Psychiatr. 2019, 25, 1218. [Google Scholar] [CrossRef] [PubMed]
- Cardose, B.M.; Sant’Anna, M..K.; Dias, V.D.; Andreazza, A.C.; Ceresér, K.M.; Kapczinski, F. The-impact of co-morbid alcohol use disorder in bipolar patients. Alcohol 2008, 42, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Schuckit, M.A.; Hesselbrock, V. Alcohol dependence and anxiety disorders: What is the relationship. Am. J. Psychiatry 1994, 151, 1723–1724. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.M.; Cleary, M.; Sitharthan, T.; Hunt, G.E. Prevalence of comorbid substance use, anxiety and mood disorders in epidemiological surveys, 1990-2014: A systematic review and meta-analysis. Drug Alcohol Depend. 2015, 154, 1–13. [Google Scholar] [CrossRef]
- Burns, L.; Teesson, M. Alcohol use disorders comorbid with anxiety, depression and drug use disorders. Findings from the Australian National Survey of Mental Health and Well Being. Drug Alcohol Depend. 2002, 68, 299–307. [Google Scholar] [CrossRef]
- Smith, J.P.; Randall, C.L. Anxiety and alcohol use disorders: Comorbidity and treatment considerations. Alcohol Res. 2012, 34, 414–431. [Google Scholar]
- Davis, L.; Uezato, A.; Newell, J.M.; Frazier, E. Major depression and comorbid substance use disorders. Curr. Opin. Psychiatry 2008, 21, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Foulds, J.A.; Adamson, S.J.; Boden, J.M.; Williman, J.A.; Mulder, R.T. Depression in patients with alcohol use disorders: Systematic review and meta-analysis of outcomes for independent and substance-induced disorders. J. Affect. Disord 2015, 185, 47–59. [Google Scholar] [CrossRef]
- Mueser, K.T.; Drake, R.E.; Wallach, M.A. Dual diagnosis: A review of etiological theories. Addict. Behav. 1998, 23, 717–734. [Google Scholar] [CrossRef]
- Hintz, T.; Mann, K. Comorbidity in alcohol use disorders: Focus on mod, anxiety and personality. In Dual Diagnosis: The Evolving Conceptual Framework; Stohler, R., Rösler, W., Eds.; Karger: Basel, Switzerland, 2005; pp. 65–91. [Google Scholar]
- Stoychev, K.R. Neuroimaging studies in patients with mental disorder and co-occurring substance use disorder: Summary of findings. Front. Psychiatry 2019, 10. [Google Scholar] [CrossRef]
- Krueger, R.F. The structure of common mental disorders. Arch. Gen. Psychiatry 1999, 56, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Krueger, R.F.; McGue, M.; Iacono, W.G. The higher-order structure of common DSM mental disorders: Internalization, externalization, and their connections to personality. Personal. Individ. Differ. 2001, 30, 1245–1259. [Google Scholar] [CrossRef]
- Kendler, K.S.; Prescott, C.A.; Myers, J.; Neale, M.C. The Structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch. Gen. Psychiatry 2003, 60, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Tawa, E.A.; Hall, S.D.; Lohoff, F.W. Overview of the genetics of alcohol use disorder. Alcohol Alcohol. 2016, 51, 507–514. [Google Scholar] [CrossRef]
- Lappalained, J. Genetic basis of dual diagnosis. In Dual Diagnosis and Psychiatric Treatment; Kranzler, J.R., Tinsley, J.A., Eds.; Marcel Dekker: New York, NY, USA, 2004. [Google Scholar]
- Nurnberger, J.I., Jr.; Foroud, T.; Flury, L.; Su, J.; Meyer, E.T.; Hu, K.; Crowe, R.; Edenberg, H.; Goate, A.; Bierut, L.; et al. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am. J. Psychiatry 2001, 158, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Reich, T.; Edenberg, H.J.; Goate, A.; Williams, J.T.; Rice, J.P.; Van Eerdewegh, P.; Foroud, T.; Hesselbrock, V.; Schuckit, M.A.; Bucholz, K.; et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet. 1998, 81, 207–215. [Google Scholar] [CrossRef]
- Foroud, T.; Edenberg, H.J.; Goate, A.; Rice, J.; Flury, L.; Koller, D.L.; Bierut, L.J.; Conneally, P.M.; Nurnberger, J.I.; Bucholz, K.K.; et al. Alcoholism susceptibility loci: Confirmation studies in a replicate sample and further mapping. Alcohol Clin. Exp. Res. 2000, 24, 933–945. [Google Scholar] [CrossRef]
- Fullerton, J.; Cubin, M.; Tiwari, H.; Wang, C.; Bomhra, A.; Davidson, S.; Miller, S.; Fairburn, C.; Goodwin, G.; Neale, M.C.; et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am. J. Hum. Genet. 2003, 72, 879–890. [Google Scholar] [CrossRef][Green Version]
- Gelernter, J.; Page, G.P.; Bonvicini, K.; Woods, S.W.; Pauls, D.L.; Kruger, S. A chromosome 14 risk locus for simple phobia: Results from a genomewide linkage scan. Mol. Psychiatry 2003, 8, 71–82. [Google Scholar] [CrossRef]
- Nash, M.W.; Huezo-Diaz, P.; Williamson, R.J.; Sterne, A.; Purcell, S.; Hoda, F.; Cherny, S.S.; Abecasis, C.R.; Prince, M.; Gray, J.A.; et al. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum. Mol. Genet. 2004, 13, 2173–2182. [Google Scholar] [CrossRef][Green Version]
- Donadon, M.F.; Osório, F.L. Personality traits and psychiatric comorbidities in alcohol dependence. Braz. J. Med. Biol. Res. 2016, 49. [Google Scholar] [CrossRef]
- Duncan, L.E.; Ostacher, M.; Ballon, J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacol 2019, 44, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Burnbaum, R.; Weinberger, D.R. Pharmacological implications of emerging schizophrenia genetics. J. Clin. Psychopharmacol 2020, 40, 323–329. [Google Scholar] [CrossRef]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Bracken, M.B.; et al. Complement factor H polymorphism in age- related macular degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Palk, A.C.; Dalvie, S.; de Vries, J.; Martin, A.R.; Stein, D.J. Potential use of clinical polygenic risk scores in psychiatry—ethical implications and communicating high polygenic risk. Philos. Ethics Humanit. Med. 2019, 14. [Google Scholar] [CrossRef]
- Coleman, J.R.I.; Gaspar, H.A.; Bryois, J. The genetics of the mood disorder spectrum: Genome-wide association analyses of over 185,000 cases and 439,000 controls. Biol. Psychiatry 2020, 88, 168–184. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Terzlaf, J.M.; Alk, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Saunders, J.B.; Aasland, O.G.; Babor, T.F.; de la Fuente, J.R.; Grant, M. Development of the alcohol use disorders identification test (AUDIT): WHO collaborative project on early detection of persons with harmful alcohol consumption—II. Addiction 1993, 88, 791–804. [Google Scholar] [CrossRef]
- Ripke, S.; Wray, N.R.; Lewis, C.M.; Hamilton, S.P.; Weissman, M.M.; Breen, G.; Byrne, E.M.; Blackwood, D.H.R.; Boomsma, D.I.; Cichon, S.; et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 2013, 18, 497–511. [Google Scholar] [CrossRef]
- Wray, N.R.; Ripke, S.; Mettheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef]
- Sklar, P.; Ripke, S.; Scott, L.J.; Andreassen, O.A.; Cichon, S.; Craddock, N.; Edenberg, H.J., Jr.; Nurnberger, J.I.; Rietschel, M.; Blackwood, D.; et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011, 43, 977–983. [Google Scholar] [CrossRef]
- Lydall, J.; Bass, N.J.; McQuillin, A.; Lawrence, J.; Anjorin, A.; Kandaswamy, R.; Pereira, A.; Guerrini, I.; Curtis, D.; Vine, A.E.; et al. Confirmation of prior evidence of genetic susceptibility to alcoholism in a genome-wide association study of comorbid alcoholism and bipolar disorder. Psychiatr. Genet. 2011, 21, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Drgon, T.; Liu, Q.R.; Walther, D.; Edenberg, H.; Rice, J.; Foroud, T.; Uhl, G.R. Pooled association genome scanning for alcohol dependence using 104 268 SNPs: Validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141B, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Kerner, B.; Lambert, C.G.; Muthén, B.O. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS ONE 2011, 6, e28477. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.C.; Aliev, F.; Bierut, L.J.; Bucholz, K.K.; Edenberg, H.; Hesselbrock, V.; Kramer, J.; Kuperman, S.; Nurnberger, J.I.; Schuckit, M.A.; et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr. Genet. 2012, 22, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Sharp, S.I.; McQuillin, A.; Marks, M.; Hunt, S.P.; Stanford, S.C.; Lydall, G.J.; Morgan, M.Y.; Asherson, P.; Curtis, D.; Gurling, H.M.D. Genetic association of the Tachykinin Receptor 1 TACR1 gene in bipolar disorder, attention deficit hyperactivity disorder and the alcohol dependence syndrome. Am. J. Med. Genet.Part B Neuropsychiatr. Genet. 2014, 165B, 373–380. [Google Scholar] [CrossRef]
- Levey, D.F.; Le-Niculescu, H.; Frank, J.; Ayalew, M.; Jain, N.; Kirlin, B.; Learman, R.; Winiger, E.; Rodd, Z.; Shekhar, A.; et al. Genetic risk prediction and neurobiological understanding of alcoholism. Transl. Psychiatry 2014, 4, e391. [Google Scholar] [CrossRef]
- Treutlein, J.; Cichon, S.; Ridinger, M.; Wodarz, N.; Soyka, M.; Zill, P.; Maier, W.; Moessner, R.; Gaebel, W.; Dahmen, N.; et al. Genome-wide association study of alcohol dependence. Arch. Gen. Psychiatry 2009, 66, 773–784. [Google Scholar] [CrossRef]
- Frank, J.; Cichon, S.; Treutlein, J.; Ridinger, M.; Mattheisen, M.; Hoffmann, P.; Herms, S.; Wodarz, N.; Soyka, M.; Zill, P.; et al. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict. Biol 2012, 17, 171–180. [Google Scholar] [CrossRef]
- Patel, S.D.; Le-Niculescu, H.; Koller, D.L.; Green, S.D.; Lahiri, D.K.; McMahon, F.J.; Nurnberger, J.I., Jr.; Niculescu, A.B., III. Coming to grips with complex disorders: Genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am. J. Med. Genet. 2010, 153B, 850–877. [Google Scholar] [CrossRef] [PubMed]
- Le-Niculescu, H.; Case, N.J.; Hulvershorn, L.; Patel, S.D.; Bowker, D.; Gupta, J.; Bell, R.; Edenberg, H.J.; Tsuang, M.T.; Kuczenski, R.; et al. Convergent functional genomic studies of omega-3 fatty acids in stress reactivity, bipolar disorder and alcoholism. Transl. Psychiatry 2011, 1, e4. [Google Scholar] [CrossRef]
- Carey, C.E.; Agrawal, A.; Bucholz, K.K.; Hartz, S.M.; Lynskey, M.T.; Nelson, E.C.; Bierut, L.J.; Bogdan, R. Associations between polygenic risk for psychiatric disorders and substance involvement. Front. Genet. 2016, 7, 149. [Google Scholar] [CrossRef]
- Bierut, L.J.; Agrawal, A.; Bucholz, K.K.; Doheny, K.F.; Laurie, C.; Pugh, E.; Fisher, S.; Fox, L.; Howells, L.; Bertelsen, S.; et al. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. USA 2010, 107, 5082–5087. [Google Scholar] [CrossRef]
- Bierut, L.J.; Madden, P.A.; Breslau, N.; Johnson, E.O.; Hatsukami, D.; Pomerleau, O.F.; Swan, G.E.; Rutter, J.; Bertelsen, S.; Fox, L.; et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007, 16, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Bierut, L.J.; Strickland, J.R.; Thompson, J.R.; Afful, S.E.; Cottler, L.B. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008, 95, 14–22. [Google Scholar] [CrossRef]
- Andersen, A.M.; Pietrzak, R.H.; Kranzler, H.R.; Ma, L.; Zhou, H.; Liu, X.; Kramer, J.; Kuperman, S.; Edenberg, H.J.; Rice, J.O.; et al. Polygenic scores for major depressive disorder and risk of alcohol dependence. JAMA Psychiatry 2017, 74, 1153–1160. [Google Scholar] [CrossRef]
- Gelernter, J.; Sherva, R.; Koesterer, R.; Almasy, L.; Zhao, H.; Kranzler, H.R.; Farrer, L. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol. Psychiatry 2014, 19, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Fuehrlein, B.S.; Mota, N.; Arias, A.J.; Trevisan, L.A.; Kachadouran, L.K.; Krystal, J.H.; Southwick, S.M.; Pietrzak, T.H. The burden of alcohol use disorders in US military veterans: Results from the National Health and Resilience in Veterans Study. Addiction 2016, 111, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Polimanti, R.; Yang, B.; Wang, Q.; Shizhong, H.; Sherva, R.; Nuñez, Y.Z.; Zhao, H.; Farrer, L.A.; Kranzler, H.R.; et al. Genetic risk variants associated with comorbid alcohol dependence and major depression. JAMA Psychiatry 2017, 74, 1234–1241. [Google Scholar] [CrossRef]
- Okbay, A.; Baselmans, B.; De Neve, J.E.; Turley, P.; Nivard, M.G.; Fontana, M.A.; Meddens, F.W.; Linner, R.K.; Rietveld, C.A.; Derringer, J.; et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 2016, 48, 624–633. [Google Scholar] [CrossRef]
- Reginsson, G.R.; Inganson, A.; Euesden, J.; Bjornsdottir, G.; Olafsson, S.; Sigurdson, E.; Oskarsson, H.; Tyrfingsson, T.; Runarsodottir, V.; Hansdottir, I.; et al. Polygenic risk scores for schizophrenia and bipolar disorder associate with addiction. Addict. Biol. 2018, 23, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Muench, C.; Schwandt, M.; Jung, J.; Cortes, C.R.; Momenan, R.; Lohoff, F.W. The major depressive disorder GWAS-supported variant rs10514299 in TMEM161B-MEF2C predicts putamen activation during reward processing in alcohol dependence. Transl. Psychiatry 2018, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Otowa, T.; Hek, K.; Lee, M.; Byrne, E.M.; Mirza, S.S.; Nivard, M.G.; Bigdeli, T.; Aggen, S.H.; Adkins, D.; Wolen, A.; et al. Metaanalysis of genome-wide association studies of anxiety disorders. Mol. Psychiatry 2016, 21, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Foo, J.C.; Streit, F.; Treutlein, J.; Ripke, S.; Witt, S.H.; Strohmaier, J.; Degenhardt, F.; Forstner, A.J.; Hoffmann, P.; Soyka, M.; et al. Shared genetic etiology between alcohol dependence and major depressive disorder. Psychiatr. Genet. 2018, 28, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Walters, R.K.; Polimanti, R.; Johnson, E.C.; McClintick, J.N.; Adams, M.J.; Adkins, A.E.; Aliev, F.; Bacanu, S.A.; Batzler, A.; Bertelsen, S.; et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 2018, 21, 1656–1669. [Google Scholar] [CrossRef] [PubMed]
- Polimanti, R.; Peterson, R.E.; Ong, J.S.; MacGregor, S.; Edwards, A.; Clarke, T.K.; Frank, J.; Gerring, Z.; Gillespie, N.A.; Lind, P.A.; et al. Evidence of causal effect of major depression on alcohol dependence: Findings from the psychiatric genomics consortium. Psychol. Med. 2019, 49, 1218–1226. [Google Scholar] [CrossRef]
- Allen, N.; Sudlow, C.; Downey, P.; Peakman, T.; Danesh, J.; Elliott, P.; Callacher, J.; Green, J.; Matthews, P.; Pell, J.; et al. UK Biobank: Current status and what it means for epidemiology. Health Policy Technol. 2012, 1, 123–126. [Google Scholar] [CrossRef]
- Martínez-Magaña, J.J.; Gonzalez-Castro, T.B.; Genís-Mendoza, A.D.; Tovilla-Zárate, C.A.; Juárez-Rojop, I.E.; Saucedo-Uribe, E.; Rodriguez-Mayoral, O.; Lanzagorta, N.; Escamilla, M.; Macías-Kauffer, L.; et al. Exploratory analysis of polygenic risk scores for psychiatric disorders: Applied to dual diagnosis. Rev. Invest. Clin. 2019, 71, 321–329. [Google Scholar] [CrossRef]
- Colbert, S.M.C.; Funkhouser, S.A.; Johnson, E.C.; Hoeffer, C.; Ehringer, M.A.; Evans, L.M. Differential shared genetic influences on anxiety with problematic alcohol use compared to alcohol consumption. MedRxiv 2020. [Google Scholar] [CrossRef]
- Purves, K.L.; Coleman, J.R.I.; Meier, S.M.; Rayner, C.; Davis, K.A.S.; Cheesman, R.; Bækvad-Hansen, M.; Børglum, A.D.; Cho, S.W.; Deckert, J.J.; et al. A major role for common genetic variation in anxiety disorders. Mol. Psychiatry 2020, 25, 3292–3303. [Google Scholar] [CrossRef]
- Sanchez-Roige, S.; Palmer, A.A.; Fontanillas, P.; Elson, S.L.; Adams, M.J.; Howard, D.M.; Mclntosh, A.M.; Clarke, T.K. Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am. J. Psychiatry 2019, 176, 107–118. [Google Scholar] [CrossRef]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Vrieze, S. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019, 51, 237–244. [Google Scholar] [CrossRef]
- Johnson, C.; Drgon, T.; McMahon, F.J.; Uhl, G.R. Convergent genome wide association results for bipolar disorder and substance dependence. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009, 150B, 182–190. [Google Scholar] [CrossRef]
- Edenberg, H.J.; Xuei, X.; Chen, H.; Tian, H.; Wetherill, L.F.; Dick, D.M.; Almasy, L.; Bierut, L.; Bucholz, K.K.; Goate, A.; et al. Association of alcohol dehydrogenase genes with alcohol dependence: A comprehensive analysis. Hum. Mol. Genet. 2006, 15, 1539–1549. [Google Scholar] [CrossRef]
- Enoch, M.A.; Hodgkinson, C.A.; Yuan, Q.; Shen, P.H.; Goldman, D.; Roy, A. The influence of GABRA2, childhood trauma, and their interaction on alcohol, heroin, and cocaine dependence. Biol. Psychiatry 2010, 67, 20–27. [Google Scholar] [CrossRef]
- Cook, C.; Gurling, H. The D2 dopamine receptor gene and alcoholism: A genetic effect on the liability of alcoholism. J. R. Soc. Med. 1994, 87, 400–402. [Google Scholar] [PubMed]
- Joe, K.H.; Kim, D.J.; Park, B.L.; Yoon, S.; Lee, H.K.; Kim, T.S.; Cheon, Y.H.; Gwon, D.H.; Cho, S.N.; Lee, H.W.; et al. Genetic association of DRD2 polymorphisms with anxiety scores among alcohol-dependent patients. Biochem. Biophys. Res. Commun. 2008, 371, 591–595. [Google Scholar] [CrossRef]
- Howard, D.M.; Adams, M.J.; Clarke, T.K.; Hafferty, J.D.; Gibson, J.; Shiali, M.; McIntosh, A.M. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci 2019, 22, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Procopio, D.O.; Saba, L.M.; Walter, H.; Lesch, O.; Skala, K.; Schlaff, G.; Tabakoff, B. Genetic markers of comorbid depression and alcoholism in women. Alcohol Clin. Exp. Res. 2013, 37, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Lesch, O.M.; Walter, H. Subtypes of alcoholism and their role in therapy. Alcohol Alcohol Suppl 1996, 31, 63–67. [Google Scholar] [CrossRef]
- Edenberg, H.D.; Koller, D.L.; Xiaoling, X.; Wetherill, L.; McClintick, J.N.; Almasy, L.; Bierut, L.J.; Bucholz, K.K.; Goate, A.; Aliev, F.; et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin. Exp. Res. 2010, 34, 840–852. [Google Scholar] [CrossRef]
- Mosheva, M.; Serretti, A.; Stukalin, Y.; Fabbri, C.; Hagin, M.; Horev, S.; Mantovani, V.; Bin, S.; Matticcio, A.; Nivoli, A.; et al. Association between CANCA1C Gene rs1034936 polymorphism and alcohol dependence in bipolar disorder. J. Affect. Disord. 2019, 261, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, K.; Almasy, L.; Knowles, E.E.M.; Kent, J.W.; Curran, J.E.; Dyer, T.D.; Göring, H.H.H.; Olvera, R.L.; Fox, P.T.; Pearlson, G.D.; et al. Genome-wide significant loci for addiction and anxiety. Eur. Psychiatry 2016, 36, 47–54. [Google Scholar] [CrossRef]
- Lionel, A.C.; Tammimies, K.; Vaags, A.K.; Rosenfeld, J.A.; Ahn, J.W.; Merico, D.; Noor, A.; Runke, C.K.; Pillalamirri, V.K.; Carter, M.T.; et al. Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum. Mol. Genet. 2014, 23, 2752–2768. [Google Scholar] [CrossRef]
- Locke, M.; Tinsley, C.L.; Benson, M.A.; Blake, D.J. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009, 18, 2344–2358. [Google Scholar] [CrossRef]
- Papaleo, F.; Weinberger, D.R. Dysbindin and schizophrenia: It’s dopamine and glutamate all over again. Biol. Psychiatry 2011, 69, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Khantzian, E.J. The self-medication hypothesis of addictive disorders. Am. J. Psychiatry 1985, 142, 1959–1964. [Google Scholar] [CrossRef]
- Schumann, G.; Coin, L.J.; Lourdusamy, A.; Charoen, P.; Berger, K.H.; Stacey, D.; Desrivières, S.; Aliev, F.; Khan, A.A.; Amin, N.; et al. Genome-wide association and genetic functional studies identidy autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. USA 2011, 108, 7119–7124. [Google Scholar] [CrossRef]
- Hori, K.; Nagai, T.; Shan, W.; Sakamoto, A.; Abe, M.; Yamazaki, M.; Sakimura, K.; Yamada, K.; Hoshini, M. Heterozygous disruption of autism susceptibility candidate 2 causes impaired emotional control and cognitive memory. PLoS ONE 2015, 10, e0145979. [Google Scholar] [CrossRef]
- Zhou, H.; Cheng, Z.; Bass, N.; Krystal, J.H.; Farrer, A.; Kranzler, H.R.; Gelernter, J. Genome-wide association study identifies glutamate ionotropic receptor GRIA4 as a risk gene for comorbid nicotine dependence and major depression. Transl. Psychiatry 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Enoch, M.A.; Rosser, A.A.; Zhou, Z.; Mash, D.C.; Yuan, Q.; Goldman, D. Expression of glutamatergic genes in healthy humans across 16 brain regions; altered expression in the hippocampus after chronic exposure to alcohol or cocaine. Genes Brain Behav. 2014, 13, 758–768. [Google Scholar] [CrossRef]
- Di Nicola, M.; Moccia, L.; Ferri, V.L.; Panaccione, I.; Janiri, L. Alcoholism in bipolar disorder: An overview of epidemiology, common pathogenic pathways, course of disease, and implications for treatment. In Neuroscience of Alcohol; Preedy, V.R., Ed.; Academic Press: London, UK, 2019; pp. 363–371. [Google Scholar] [CrossRef]
- Jensen, K.P.; Stein, M.B.; Kranzler, H.R.; Yang, B.Z.; Farrer, L.A.; Gelernter, J. The α-endommanosidase gene (MANEA) is associated with panic disorder and social anxiety disorder. Transl. Psychiatry 2014, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sipilä, T.; Kananen, L.; Greco, D.; Donner, J.; Silander, K.; Terwilliger, J.D.; Auvinen, P.; Peltonen, L.; Lönnqvist, J.; Pirkola, S.; et al. An association analysis of circadian genes in anxiety disorders. Biol. Psychiatry 2010, 67, 1163–1170. [Google Scholar] [CrossRef]
- Spanagel, R.; Pendyala, G.; Abarca, C.; Zghoul, T.; Sanchis-Segura, C.; Magnone, M.C.; Lascorz, J.; Depner, M.; Holzberg, D.; Soyka, M.; et al. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat. Med. 2005, 11, 11–35. [Google Scholar] [CrossRef]
- Le-Niculescu, H.; McFarland, M.J.; Ogden, C.A.; Balaraman, Y.; Patel, S.; Tan, J.; Rodd, Z.A.; Paulus, M.; Geyer, M.A.; Edenberg, H.J.; et al. Phenomic, convergent functional genomic, and biomarker studies in a stress-reactive genetic animal model of bipolar disorder and co-morbid alcoholism. Am. J. Med. Genet. B Neuropsychiatr Genet. 2008, 147B, 134–166. [Google Scholar] [CrossRef]
- Fullerton, J.M.; Nurnberger, J.L. Polygenic risk scores in psychiatry: Will they be useful for clinicians? F1000Research 2019, 8, F1000 Faculty Rev-1293. [Google Scholar] [CrossRef]
- Birkett, J.T.; Arranz, M.J.; Munro, J.; Osbourn, S.; Kerwin, R.W.; Collier, D.A. Association analysis of the 5-HT5A gene in depression, psychosis and antipsychotic response. Neuro Rep. 2000, 11, 2017–2020. [Google Scholar] [CrossRef] [PubMed]
- Yosifova, A.; Mushiroda, T.; Stoianov, D.; Vazharova, R.; Dimova, I.; Karachanak, S.; Zaharieva, I.; Milanova, V.; Madjirova, N.; Gerdjikov, I.; et al. Case-control association study of 65 candidate genes revealed a possible association of a SNP of HTR5A to be a factor susceptible to bipolar disease in Bulgarian population. J. Affect. Disord. 2009, 117, 87–97. [Google Scholar] [CrossRef]
| Study | Sample | Identified High-Risk Polymorphisms/PRS Associations | Neurobiological Underpinnings | Comments |
|---|---|---|---|---|
| Lydall et al. 2011 [46] | 506 bipolar I disorder (BPD-I) cases (m = 193) from the University College of London cohort 1 (UCL1) and 510 controls (m = 217). Cases were of English (Caucasian), Irish, Scots and Welsh ancestry. Two phenotypes were defined: ICD-10/DSM-III-R bipolar disorder + RDC 1 alcoholism (BPALC)—143 cases (m = 80); Bipolar disorder without alcoholism (NABPD)—363 cases (m = 113). | Suggestive significance (p < 1 × 10−1)) was detected for the following SNPs, located in or near to genes previously implicated in alcoholism: rs429065 (16q22, p = 1.03 × 10−4) in the region of CDH11 (cadherin 11 gene); rs3130159 (chr. 6p21.3, p = 2.83 × 10−3) within COL11A2 (collagen type 11 a2 gene); rs17113138 (5q33.1, p = 1.43 × 10−2) within NMUR2 (neuromedin U receptor 2 gene); rs7013323 (8p21.3, p = 7.99 × 10−3) within XPO7 (exportin 7 gene); rs2256569 (5p15.31, p = 2.11 × 10−4)—SEMA5A (semaphorin-associated protein 5A gene). | Cadherin11—belongs to a group of transmembrane proteins that mediate Ca++dependent cell–cell adhesion and the generation of synaptic complexity in the developing brain, implicated in mnemonic processes, addictions, and BPD. COL11A2—encoding one of the two chains of XI collagen; implicated in various facial and skeleton bone dysplasia syndromes as well as Mendelian inherited sensorineural deafness; NMUR2—belongs to the G-protein coupled receptor 1 family and is expressed in guts and CNS (hypothalamus). It binds to the neuropeptide U. The receptor plays a role in food intake and body weight. XPO7 protein—mediates the nuclear export of proteins with broad substrate specificity. Involved in alcoholism according to the pooled COGA genome associated data [47]; SEMA5A—a member of the semaphorins family of membrane proteins involved in axonal guidance during neural development. Associated with autism susceptibility. | Most significant SNPs associations with BPALC phenotype were within or near genes involved in cell adhesion, differentiation and regulation, neurotransmitter pathways and ion function, enzymatic activity, cellular messengers (second messengers), connective tissue. Association between these SNPs and the BPALC cases, but not with NABPD cases suggests genetic effects on alcoholism independent of bipolar affective disorder. Genes of the GABA system (e.g., GABA receptor type 2), which are among the most replicated in alcoholism, were not determined as being associated with BPALC phenotype, indicating either genetic heterogeneity of alcoholism, or the possibility that alcoholism in BPD is mediated by different pathways. Limitations: Small sample for a GWAS. |
| Kerner et al. 2011 [48] | 1000 EA 2 subjects (m = 499) with BPD-I (DSM-IV) and 1034 controls (m = 532). In BPD sample, 250 patients (group 1) had lifetime alcohol dependence (AD) with or without lifetime substance abuse/dependence (including nicotine); 40% of another group of 270 patients with BPD with psychotic features (group 2) had a lifetime alcohol abuse but not AD. | The SNP rs2727943 (3p26.3, p = 3.36 × 10−8) was associated with OR of 4.9 for having BD-I with comorbid alcohol dependence (group 1) phenotype. This polymorphism is located between the genes contactin-4 precursor (BIG-2) and neural adhesion molecule contactin 6 (CNTN6). In group 2, statistical significance on genome-wide level was detected for rs1039002 (6q27.5, p = 1.7 × 10−8) and rs12563333 (1q41, p = 5.9 × 10−8). Besides, two SNPs neared significance: rs9493867 (6q23.2, p-value from 1.0 × 10−7 under recessive model to 9.0 × 10−8 under dominant) within the gene encoding serine/threonine kinase (Skg1); and rs13220542 (6q15, p = 9.0 × 10−8 under dominant model) located 3′ to the gene coding mitogen-activated protein kinase, kinase, kinase 7 (MAP3K7). | The high-risk SNP is located in a region that is deleted in individuals with autistic features. Proteins coded by BIG-2 and CNTN6 might play a role in the formation of axon connections in the developing brain. rs1039002 is located in transcribed genomic sequence with unknown function. The nearest known gene is phosphodiesterase 1 (PDE10A) which is involved in the elimination of intracellular cAMP and cGMP signaling molecules. Inhibitors of the PDE10A have shown therapeutic potential in Parkinson’s and Huntington’s disease, addiction, and OCD and are being tested in clinical trials. Rs12563333 is located in a transcribed sequence immediately upstream of the gene MAP/microtubule-affinity regulating kinase 1 (MARK1). MARK1 phosphorylates microtubules associated proteins and is involved in synaptic plasticity and dendritic trafficking. The two SNPs closing genome- wide significance are located in genes involved in response to stress through K, Na, and Cl channels (Skg1) and activation of protein kinases such as MAPK8 and MAP2K4 (MAP3K7). | The study distinguished three distinct profiles of comorbidity in the BP-1 sample with two of them significantly associated with specific SNP/SNPs: a group with comorbid psychosis and substance abuse (including alcohol abuse but no alcohol dependence); a group with comorbid alcohol dependence but also high lifetime prevalence of comorbid PD; and a group with a very low rate of co-morbid conditions. This suggests that phenotype heterogeneity in BPD might indicate genetic heterogeneity. The SNPs close to genome-wide significance are in genes implicated in stress response and warrant further investigation in samples with BD comorbid with SUD. Since the associated variants were rare, future studies applying re-sequencing of these chromosomal regions in BP patients could be more appropriate for replication. Limitations: Small sample for a GWAS. |
| Edwards et al. 2012 [49] | 467 cases with DSM-IV AD and major depression (MDD) phenotype (m = 287) and 407 unaffected controls (m = 132). Cases were drawn from the COGA study sample [29] and were from EA and AA 3 descent (so were controls). | No marker met genome-wide significance criteria (5 × 10−8); 10 SNPs had p values < 1 × 10−5 and 7 of them fell into the regions of the known genes: OXTR (oxytocin receptor gene), FAF1 (Fas-associated factor 1), OPA3 (Optic atrophy 3), EFHA2 (EF-hand domain family, member 2), FHIT (fragile histidine triad gene), WDR7 (WD repeat domain 7), SPATA13 (spermatogenesis associated 13). A number of SNPs with p- value < 1 × 10−3 were detected in glutamate receptor genes (GRIN2A, GRIN2C, and GRID1) which have been previously associated with SCH, MDD, and addiction; as well as within genes previously associated with depression, AD, or other addictions (CDH13, CSMD2, and HTR1B), | FAF1, OPA, EFHA2, and WDR7 genes encode protein products engaged in apoptosis (among their other functions) FHIT’s coded enzyme is involved in purine metabolism, but also in protection against DNA damage SPATA13 encodes a protein involved in cell migration, adhesion, assembly and disassembly. CDH13—a member of the cadherin family of cell adhesion molecules, impacts GABA functioning and is a risk gene for ADHD, SUD, MDD, and violent behavior; CSMD2—codes a synaptic transmembrane protein involved in the development and maintenance of dendrites and synapses that has been linked to schizophrenia and autistic disorders by GWAS. HTR1B—codes 5-HT receptor 1B receptor associated with OCD, personality disorders, and schizophrenia. | The degree of overlap of significant SNPs between a comorbid phenotype (AD-MDD) and an AD-only phenotype is modest suggesting that comorbid phenotype is partially influenced by genetic variants that do not affect AD alone. Limitations: small sample size for GWAS; >50% of cases have not met DSM-IV criteria for independent MDD, i.e., depressive symptoms in them have occurred under the influence of alcohol or drugs |
| Sharp et al. (2014) [50] | The sample consisted of 2096 patients and 1056 controls. Patients were distributed as follows: 506 BPD-I cases from UCL1; 593 cases (m = 219) from the University College of London cohort 2 (UCL2), of which 409 were with BPD-I and 184 with BPD-II; 997 AD syndrome cases from the University College of London ADS sample (UCL ADS) part of the UK-COGA project; 35 cases of ADHD 4Cases were of English (Caucasian), Irish, Scots, and Welsh ancestry. Control subjects were comprised of 672 screened individuals without mental disorder or family history for schizophrenia, AD, BPALC, and 384 unscreened ones. | Two SNPs in tachykinin receptor 1 gene (TACR1, 2p12) were significantly associated with BPD cases in comparison with screened controls—rs3771829 (p = 0.002, OR 1.57) and rs3771833 (p = 0.004, OR 1.43). However, neither of the two were associated with BPD in the UCL2 sample alone. In comparison with controls, rs3771829 was significantly associated with BPD (UCL1 and UCL2 combined, p = 9.0 × 10−8 under dominant model), ADS (p = 2.0 × 10−3) and BPALC (p = 6.0 × 10−4). DNA sequencing in selected cases of BPD and ADHD with inherited TACR1-susceptibility haplotypes determined 19 SNPs in different regions of TACR1 that increase vulnerability to BPD, ADS, ADHD, and BPALC. The association with TACR1 and BPAD, ADS, and ADHD suggests a shared molecular pathophysiology between these disorders. | Neurokinin 1 receptors (NK1R) encoded by TACR1 are abundantly expressed throughout brain regions driving reward and reinforcement. The binding density of NK1R is highest in the locus coeruleus involved in mood regulation and response to stress. Inactivation of NK1Rs critically modulates alcohol reward and escalation, supporting a direct role of NK1R in the regulation of alcohol intake and the development of alcohol dependence. NK1Rs are an attractive molecular target for the treatment of alcohol use disorders but also depression and anxiety. Trials of the efficacy of NK1R antagonists in ADS are currently underway. | The lack of association of the two top marker SNPs with BPD in UCL2 sample may reflect both the heterogeneity of BPD susceptibility genes even in single ancestrally originating cases, as well as the presence of low frequency disease alleles. Differences of association of rs3771829 and rs3771833 was stronger for BPALC only compared to screened controls than for BPD-total compared to screened controls. Therefore, it is likely that the comorbid ADS in BPD cohort is driving the association, i.e., NK1Rs are more strongly implicated in the neurobiology of alcohol use disorders than in BPD. Limitations: Small sample for a GWAS. |
| Levey et al. 2014 [51] | 7948 subjects (4519 patients and 3429 controls). Patients were distributed as follows: 1151 men from German-Caucasian descent with AD [52,53]; 2768 patients (m = 1687) from EA (n = 1273) and AA (n = 1495) origin with AD; 600 patients (m = 366) from EA (324) and AA (n = 276) origin with alcohol abuse. Controls were distributed as follows: 2168 subjects form German-Caucasian descent (m = 939); 1261 subjects (m = 475) from EA (388) and AA (873) descent. | Authors used a translational Convergent Functional Genomics (CFG) approach to discover genes involved in alcoholism by integration of GWAS data with other genetic and gene-expression data from human and animal model studies. Top 11 candidate genes for alcoholism detected by this study (p = 0.041) were explored for the degree of overlap with risk genes for BPD and anxiety disorders in previous studies of the same group with a similar design [54,55]. Besides, the association of Genetic Risk Predictive Panel for BPD (i.e., list of top risk SNPs in 56 genes involved in BPD) identified by the same authors in a previous study [54] was tested in AD, AA, and Control samples of the current study. Results: SNPs in SNCA (rs17015888) and DRD2 were shared among anxiety disorder and alcoholism samples while GNAI1, GRM3 (rs17160519 to rs4236502), and MBP genes showed involvement in BPD and alcoholism. Genetic risk prediction score for BP showed increased genetic load for bipolar disorder in both alcohol dependence (p = 9.94 × 10−8) and alcohol abuse (p = 1.18 × 10−4). | SNCA (synuclein alpha), a pre-synaptic chaperone, has been reported previously as being involved in modulating brain plasticity and neurogenesis, as well as neurotransmission, primarily as a brake. On the pathological side, low levels of SNCA might offer less protection against oxidative stress, whereas high levels of SNCA may have a role in neurodegenerative diseases, like Parkinson’s disease. SNCA has been described as a susceptibility gene for alcohol cravings and response to alcohol cues. DRD2 (dopamine 2 receptor gene) has shown reduced expression in the brains of alcoholics and one possible explanation for this, bridging the common role of this receptor in AUD and BPD, is that both conditions include hyperdopaminergic state which drives individuals to hedonistic activities and leads to homeostatic downregulation of their DRD2 receptors. An alternative hypothesis sees lower levels of dopamine receptors as a reflection of reduced dopaminergic signaling and anhedonia, leading individuals to overcompensate by alcohol and drug abuse. GRM3 belongs to the metabotropic glutamate receptors family (G protein-coupled receptors), i.e., it is heavily involved in neurotransmitter signaling. GRM3 has been exclusively associated with BPD so far. MBP (Myelin Basic Protein gene) encodes a major constituent of the myelin sheath of oligodendrocytes and Schwann cells GNAI1 (Guanine Nucleotide-Binding Protein G(I) Subunit Alpha-1 gene) encodes a protein that is part of a complex that responds to beta-adrenergic signals by inhibiting adenylate cyclase. | The GWAS study on which discovery was based contained males as probands and males and females as controls. Therefore, it is possible that some of the nominally significant SNPs detected have to do with gender differences rather than with alcoholism per se, or at least, are limited to male alcoholism. Stratification across gender and ethnicities may have also been a confounding factor in US samples. Possible ethnicity differences in alleles, genes, and the consequent neurobiology need to be explored in further larger sample studies, taking into account environmental and cultural factors. Limitations: Small sample for a GWAS. |
| Carey et al. 2016 [56] | 1160 cases and 1413 controls (f = 56%) included in SAGE [57] and sampled from three previous studies—COGA [29], COGEND [58], and FSCD [59]. Cases were of non-Hispanic EA ancestry. In addition to meeting DSM-IV AD criteria, they often met criteria for cocaine, cannabis, and opioid dependence. A measure of general substance involvement (GENSUB) was generated by factor analysis of the individual substance involvement measures (types of substances and frequency of use). Associations between PGC-PRS-MDD1 and PGC-PRS-BPD1 and involvement in AD were tested. PRS-MDD1 includes >200 SNPs related to >180 genes associated with synaptic function and neurotransmission and especially expressed in prefrontal brain areas. BP-PRS1 includes SNPs within or near genes implicated in cell adhesion and migration as well as coding of calcium and other ion channels, neurotransmitter receptors, and synaptic components. | MDD-PRS/Alcohol Dependence: suggestive significance (p < 0.0001, OR 1.23, 95%CI) was found for associated MDD-PRS and severe alcohol dependence (6–7 dependence symptoms), supporting shared genetic liability to MDD and AD. Current MDD-PRS explain roughly 1% of the variance in general substance involvement (GENSUB). BPD-PRS/Alcohol Dependence: there was evidence for a dose-dependent relationship between BPD-PRS and an increasing number of alcohol dependence symptoms among regular drinkers with at least one symptom of dependence. Association of BPD-PRS with GENSUB (i.e., involvement in multiple substances) was much stronger than that for alcohol alone. | MDD/Alcohol Dependence: together with data from previous GWAS showing significant overlapping regions/variants contributing specifically to MDD alone, MDD with a comorbid SUD, or a combined MDD and SUD phenotype only, these results suggest that relationships between MDD and alcohol and MDD and other substances (cocaine) are substance-specific. The overall association with general substance involvement liability may be reflective of similar cognitive mechanisms (e.g., impulsivity, emotion dysregulation, sensation- seeking) that are thought to broadly underlie both BPD and substance use disorders. Such a mechanism fits well with the hypothesis for a genetic basis of the BPD-SUD comorbidity supported by many studies. | Limitations: 1. Small sample for a GWAS. Nominal associations may strengthen with larger discovery samples (e.g., samples from which PRS are extracted), as well as larger target samples. 2. While the study confirms that shared genetic architecture contributes to mood disorders and substance use disorders, it does not reveal specific biological (e.g., reward-related neural responsiveness, epigenetically driven gene expression changes), psychological (e.g., anhedonia, impulsivity), and/or experiential (e.g., early life stress, peer group pressure) mechanisms through which risk is manifested. |
| Andersen et al. 2017 [60] | 3871 DSM-IV-AD cases (m = 2551) and 3347 controls (m = 2082) from four different study samples—COGA [29], SAGE [57], Yale-Penn [61], and NHRVS [62]. Subjects were of European-American ancestry. The prevalence of MDD among AD patients in the different samples was between 19 and 35% and for controls—between 6.3 and 12.5%. Associations between PGC-MDD-PRS1 and AD were performed with the analysis corrected for age, sex, and population stratification. | A significant association was observed between MDD-PRS and AD case-control status for all four AD samples (p = 3.3 × 10−9; p value threshold = 0.4). The proportion of variance in AD explained by the MDD-PRS was small (R2 value of 0.0018 (min.) and 0.026 (max). Association remained even when recalculated MDD-PRS from GWAS-MDD samples without comorbid MDD-AD cases was used in analyses performed only for those patients from the four samples with pure AD (i.e., without MDD), providing further support for the genetic overlap between MDD and AD. No difference in the strength or significance or associations between MDD-PRS and AD status by sex was observed. | Although studies like the current one cannot, due to their design, determine the mechanisms by which shared genetic liability for MDD and AD operate, there are some suggestive possibilities that should be tested by future studies. For example, anxiety may be a significant factor linking AD and MDD via the internalizing pathway. Furthermore, broader personality traits such as neuroticism, disinhibition, and sensation seeking are potentially associated with a range of internalizing and externalizing psychiatric disorders, including comorbidity of MDD and AD. | Limitations: 1. MDD-GWAS with larger sample sizes will likely improve the predictive ability of MDD PRS and probably lead to refinement of observed associations. 2. The study included only AD case (and not other SUDs) and for this reason further studies are needed to check whether the MDD-PRS association is specific to AD or it generalizes to substance dependence broadly as suggested by existing research data. |
| Zhou et al. 2017 [63] | 7822 subjects (m = 4480) from EA (3169) and AA (4653) descent from the Yale-Penn Study [61] with lifetime DSM-IV AD and MDD diagnosis. The participants were divided into Yale-Penn 1 and Yale-Penn 2 subsamples based on the period of recruitment (between 1999 and 2015) and on the genotyping platform used. | The SNP rs139438618 at the SEMA3A (semaphorin 3A) gene locus was significantly associated with AD and MD comorbidity in AA participants in the Yale-Penn 1 (β = 0.89; p = 2.76 × 10−8) and Yale-Penn 2 (β = 0.83; p = 2.06 × 10−4) There was no significant association identified in EA participants. Analyses of PRS showed that individuals with a higher risk of neuroticism or depressive symptoms and a lower level of subjective well-being and educational attainment had a higher level of AD and MD comorbidity, while larger intracranial and smaller putamen volumes were associated with higher risks of AD and MD comorbidity. | Rs139438618 is located in the intron part of the SEMA3A gene (7q21.11), which codes the homonymous protein part of the semaphorin family. The latter consists of transmembrane and secretion proteins involved in the axonal growth and connectivity acting like chemorepulsors (inhibitors of axonal sprouts) or chemoattractants (stimulators of apical dendrites). The expression of these genes is most intensive in early fetal development in the olfactory brain and cerebral and entorhinal cortex. Previous studies have confirmed their role in schizophrenia, Alzheimer’s disease, epilepsy, and amyotrophic lateral sclerosis as well as intestinal malformations (Hirschprung disease). AD-MDD phenotype was associated with neuroticism PRS including 11 significant SNPs on chromosomes 3, 8, 9,11,15,17,18. One of them, on chromosome 8, is in the zone of MSRA and MTMR9 genes which are both expressed in CNS and code products engaged in repair of oxidatively damaged proteins (MSRA) and cell proliferation control (MTRM9). Both of them are significantly associated with depression/neuroticism and low subjective well-being according to a large GWAS (n = 170,000) [64]. In addition to that, MTRM9 has also been linked to generalized epilepsy with febrile seizures. | The rs139438618 SNP has not so far been identified as risk associated in GWAS studies with pure MDD and pure AD phenotypes, which suggests a pleiotropic effect on the level of a single gene. The presence of this SNP AA only is likely to represent a populational genetic effect. |
| Reginsson et al. 2018 [65] | 8701 cases (f = 32.7%) of alcohol use disorder (DSM-IIIR and DSM-IV) were part of a larger sample (n = 144,609) of Icelandic subjects, including 10,036 individuals admitted for in-patient addiction treatment, 35,754 smokers, and a group of patients with schizophrenia (n = 600) and BPD (n = 772). PGC-BPD1 PRS was tested for association with alcohol dependence. | Higher BPD1-PRS was associated with increased risk of alcohol use disorder (p = 1.7 × 10−9) and with earlier onset of substance use problems (including alcohol) (OR = 1.16, p = 1.9 h 10−3). Only alcohol use disorder (and not smoking and other substance use disorders) was nominally associated (OR = 1.09, R2 = 0.59%, p = 2.7 × 10−3) with BPD-PRS when including PGC-SCZ-PRS [66] as a covariate. This implies that alcoholism may share common genetic causal factors with BPD to a larger extent than smoking and other substance use disorders do. | The results support the notion of common genetic roots of the comorbidity between addiction (including alcohol addiction) and severe mental disorders such as BPD and schizophrenia, as opposed to solely being a direct consequence. | |
| Muench et al. 2018 [67] | BOLD fMRI 5 sample of 45 DSM-IV-AD cases with mean age (m = 35), 45 controls (m = 22) scanned during MID 6 task-directed on winning money or avoiding money loss. Subjects were of AA, EA, Asian, multiracial, and unknown ancestry. 12.4% of patients and 1.1% of controls had lifetime anxiety disorders, while current anxiety disorders measures were detected in 9.0% of patients and 1.1 of healthy controls. For mood disorders the corresponding numbers were 11.2% and 9% (lifetime) and 5.6% and 0% (current). NIAAA 7 sample of 1123 AD cases (m = 323), 735 controls (m = 325). NIAAA subsample of 955 subjects with lifetime AD (669 males). Subjects were of AA (1178), EA (1383), Asian (68), Multiracial (59), Native American/Alaska, Hawaiian/Pacific (15), and unknown ancestry (103). 5.8% of the patients and 0.4% of the controls met current MDD criteria, while 11.6% of patients and 26% of controls had a lifetime MDD. SAGE sample [57] with 1848 AD cases (1162 EA, 685 AA, males = 60%), and 1990 controls (1346 EA, 644 AA, males = 30 and 36%). | Suggestive significance (p = 0.09) was found for the previously associated with MDD risk variant rs10514299 within TMEM161B- MEF2C gene cluster containing Transmembrane Protein 161B gene and myocyte enhancer factor 2C gene. Carrying the minor T allele (TT/CT and not CC) was associated with a lifetime diagnosis of AD (odds ratio = 0.82, p = 0.09) in the NIAAA sample. The T allele of rs10514299 was significantly associated with greater depression symptom severity in individuals with a lifetime AD diagnosis (β = 1.25, p = 0.02) in the NIAAA sample with this finding driven by individuals of AA ancestry. | TMEM161B’s function is unclear, with gene ontology annotation related to it include nucleic acid binding. MEF2C encodes a transcription factor that has been so far associated with epilepsy and intellectual disability. Carrying the T allele in rs10514299 was associated with a significant increase in putamen activation during high and low loss anticipation in patients with AD, but with a significant decrease in the controls, indicating that the allele differentially affects this neural phenotype in AD. Hence, MDD risk variant rs10514299 in TMEM161B- MEF2C gene cluster was shown for the first time to predict neuronal correlates of reward processing in an AD phenotype implying possible eligibility of this polymorphism as a biomarker for disrupted reward processing in AD individuals. The fact that a MDD risk variant was also shown to be relevant to AD phenotype supports a potential role of the respective genetic locus in an endophenotype related to deficit of reward processing (i.e., anhedonia). | Limitations: 1. No correction for multiple comparisons was done, therefore future confirmatory analyses are needed to validate the functional relevance of rs10514299. 2. Insufficient sample size to detect firmly the likely small effect size of this SNP. In co-occurrence of anxiety disorders for example (GAD, panic disorder and phobias), studies reporting suggestively shared genetic susceptibility loci have employed much larger samples [68]. |
| Foo et al. 2018 [69] | Target sample AD: 1333 German-Caucasian male DSM-IV-AD cases and 1307 German-Caucasian controls from both sexes. A subset of the AD cases (n = 332) was recruited explicitly excluding comorbid MDD. Target sample MDD: 597 cases and 1292 controls from German-Caucasian ancestry (52,53). Discovery samples: PGC-PRS-MDD1 (8148 cases, 7955 controls); PGC-PRS-MDD2 (59,265 cases, 112,092 controls). | Significant associations were found between AD disease status and both PGC-PRS-MDD2 (p-threshold = 1.0, p = 0.00063, R2 = 0.533%) and PGC-PRS-MDD1 (p-threshold = 0.2, p = 0.00014, R2 = 0.663%) with the larger sample of PGC-MDD2 not building on additional predictive power. In the MDD target sample however, calculating PRS yielded more power with the bigger sample of the PGC-PRS-MDD2 (p-threshold = 1.0, p = 0.000038, R2 = 1.34%) versus PGC-PRS-MDD1 (p-threshold = 1.0, p = 0.0013, R2 = 0.81%). When calculating PGC-PRS-MDD2, PRS in the subsample of AD patients without comorbid MDD, significant associations were still found (p-threshold = 1.0, p = 0.042, R2 = 0.398%). | The presence of an association between AD disease status and PRS-MDD in a subsample of AD cases without comorbid MDD supports the hypothesis for a substantial genetic overlap between AD and MDD. Although PRS association studies like the present one do not, for the reason of design, possess the power to predict suggestive neurobiological pathways explaining comorbid phenotypes, the authors hypothesized that the level and risk of AD and MDD comorbidity may be linked to neuropsychiatric traits and brain volumes. | Limitations: determining shared genetic etiology in comorbid disorders is inevitably facing the problem of “enrichment” of the comorbid disorders in discovery and target samples. In the current study, there was no information regarding the AD comorbidity of the PGC-PRS-MDD2 sample. For that reason, future studies should employ rigorous phenotyping and improved characterization of samples with particular detailed assessment of comorbidity, symptomatology, and severity. |
| Walters et al. 2018 [70] | 14,904 individuals with DSM-IV-AD and 37,944 controls from 28 case-control and family-based studies conducted in USA, Europe and Australia. Data were stratified by genetic ancestry (European, n = 46,568; African, n = 6280). | Independent, genome-wide significant effects of different Aldehyde Dehydrogenase 1B (ADH1B) gene variants were identified in European (rs1229984; p = 9.8 × 10−13) and African ancestries (rs2066702; p = 2.2 × 10−9) Significant genetic correlations were observed between AD and 17 phenotypes in unrelated European samples (10,206 AD cases and 28,480 controls), including neuroticism (p = 2 × 10−6), depressive symptoms (p = 3 × 10−7), and MDD (p = 3 × 10−11). | Given the stringent criteria for patient selection in the study sample (i.e., all individuals were with confirmed AD diagnosis and not less severe forms of alcohol misuse), MDD may primarily share genetic liability with alcohol use at pathological levels and on a molecular level, pleiotropic effects may be implicated. | There is a continuing need to characterize the genetic architecture of AD in non-EU populations and test the genetic correlation between this phenotype and mood/anxiety phenotypes in individuals from non-European ancestry. Larger future samples will allow us to uncover additional pleiotropy between pathological and non-pathological alcohol use, as well as between AD and other neuropsychiatric disorders. |
| Polimanti et al. 2019 [71] | PGC-MDD2 sample (135,458 cases and 344,901 controls); PGC-AD sample 10,206 cases and 28,480 controls; UK Biobank sample [72] 428,308 individuals from white British ancestry. Four phenotypes were defined: MDD, AD, Alcohol Consumption—Frequency (AC-f), and Alcohol Consumption—Quantity (AC-q). | Linkage disequilibrium score regression and Mendelian randomization (MR) showed a positive genetic correlation between MD and AD. AC-q demonstrated a positive correlation with both AD and MD, while AC-f had a negative correlation with MDD and non-significant with AD. MR analyses confirmed the presence of pleiotropy among these four traits. However, the MD-AD results reflect a mediated pleiotropy mechanism (i.e., causal relationship) with an effect of MD on AD, while there was no evidence for reverse causation. | The study supports a causal role for genetic liability of MD on AD based on genetic datasets including thousands of individuals. | Larger AD and MD datasets will be required to confirm the study findings using genetic instruments based on genetic variants that reached the more conservative genome-wide significance (i.e., p < 5 × 10−8 for a particular SNP). |
| Martínez-Magaña et al. 2019 [73] | 192 individuals of Mexican Ancestry (125 cases and 67 controls). 72 of the cases had e lifetime DSM-IV schizophrenia (SCZ), while 53 (m = 25) were with BPD diagnosis. Of the latter, 23 had AD or alcohol abuse (in some combined with other SUD—nicotine, cocaine, cannabis, inhalants, or stimulants). Correlational testing of the variance of dual diagnosis (DD 8) phenotype explained by PGC-MDD1-PRS, PGC-BD1-PRS, and PGC-SCZ-PRS was performed, i.e., the hypothesis of whether the current PRS might correlate with a lifetime DD was checked. | PGC-MDD1-PRS showed a significant shared genetic etiology with the DD phenotype (Nagelkerke Pseudo-R2 = 0.0451, corr. p = 0.0118, n = 334 SNPs) whereas BPD-PRS did not (p = 0.1585). Patients with DD in the BD group had a higher MDD-PRS when compared to non-DD BD patients (p < 0.05). Notably, PGC-SZC-PRS [66] also demonstrated statistically significant common genetic background with DD phenotype, including DD-BPD (Pseudo-R2 = 0.0283, corr. p = 0.0118, n = 8058 SNPs), but it could not discriminate statistically DD-BD cases from non-DD-BD ones. Besides, MDD-PRS explained a higher amount of variance (4.51%) predicting placement in the DD group (for both SCZ and BPD patients) than did the SCZ:PRS (2.83%). | The study results suggest that both the MDD-PRS and the SCZ-PRS might be useful in detecting DD risk. However, when PRSs are applied to a specific diagnosis, MDD-PRS used in patients with BD is the only specific PRS which discriminates DD from non-DD cases. The shared genetic susceptibility between MDD and AD (or alcohol abuse) might drive this result given the fact that the main problem substance in the studied sample was (apart from nicotine) alcohol. | The study is one of the first approximations on how to apply psychiatric PRS in admixed populations (i.e., with ancestry different from European, European-American, or African-American). The application of PRS in different populations, with distinct admixtures and diverse phenotypes, could give more information on the use of PRS for psychiatric disorders as a translational risk prediction. Limitations: small sample size. |
| Colbert et al. 2020 [74] | >900,000 subjects (cases and controls) from five different GWAS samples with anxiety disorders [68,75], AD [70], problem alcohol use (PAU) [76], and alcohol consumption (AC) [77] phenotypes. | All anxiety phenotypes showed a significant positive genetic correlation with AUDIT-p (items of AUDIT associated with problem alcohol use) and AD (rg ≥ 0.35). However, the anxiety phenotypes were uncorrelated with AUDIT-C (part of AUDIT-related AC) and drinks per week, indicating AC was not genetically related to anxiety. In females, three significant positive genetic correlations were found between PAU and anxiety phenotypes DSM-V-like GAD 9 and any anxiety disorder. No such correlation was observed in males. 47 independent loci with significant (p < 7.60 × 10−7) local genetic covariance between pairs of traits were identified and three of them showed positive local genetic covariance between PAU and anxiety phenotypes. | One of the identified loci is at chr. 11 (11:113 105 405-113 958 177) and it contains the dopaminergic pathway gene (DRD2) which substantially moderates stress-induced alcohol consumption in mice and also influences connectivity between basal ganglia and frontal cortices. The region also contains NCAM (Neural Cell Adhesion Molecule gene), TTC12 (Tetratricopeptide Repeat Domain 12 gene) and ANKK1 (Ankyrin Repeat and Protein Kinase Domain-Containing Protein 1 gene) which, together with DRD2, form the so-called NTAD gene suggestively contributing to various psychiatric disorders as well as the comorbidity of psychiatric disorders. A locus on chromosome 9 (10 879 18811 616 822), previously not implicated in AUD, was also found to have multiple significant positive covariances between anxiety and PAU, but not AC. This locus has been previously associated with worry and neuroticism, depression, and anxiety but has not been associated with alcohol misuse anxiety comorbidity. | Genetic covariance between anxiety traits and PAU is concentrated in certain brain areas: amygdala, caudate basal ganglia and frontal cortex. These results align findings from fMRI studies pointing to the role of these regions in anxiety and alcohol use. Limitations: GWAS sample sizes are not large enough given the small variations of risk associated with identified loci. Limited panels for expression is another limitation of the study. Finally, the analyses does not identify specific mechanisms which contribute to the comorbidity of the two disorders or a causal direction. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stoychev, K.; Dilkov, D.; Naghavi, E.; Kamburova, Z. Genetic Basis of Dual Diagnosis: A Review of Genome-Wide Association Studies (GWAS) Focusing on Patients with Mood or Anxiety Disorders and Co-Occurring Alcohol-Use Disorders. Diagnostics 2021, 11, 1055. https://doi.org/10.3390/diagnostics11061055
Stoychev K, Dilkov D, Naghavi E, Kamburova Z. Genetic Basis of Dual Diagnosis: A Review of Genome-Wide Association Studies (GWAS) Focusing on Patients with Mood or Anxiety Disorders and Co-Occurring Alcohol-Use Disorders. Diagnostics. 2021; 11(6):1055. https://doi.org/10.3390/diagnostics11061055
Chicago/Turabian StyleStoychev, Kaloyan, Dancho Dilkov, Elahe Naghavi, and Zornitsa Kamburova. 2021. "Genetic Basis of Dual Diagnosis: A Review of Genome-Wide Association Studies (GWAS) Focusing on Patients with Mood or Anxiety Disorders and Co-Occurring Alcohol-Use Disorders" Diagnostics 11, no. 6: 1055. https://doi.org/10.3390/diagnostics11061055
APA StyleStoychev, K., Dilkov, D., Naghavi, E., & Kamburova, Z. (2021). Genetic Basis of Dual Diagnosis: A Review of Genome-Wide Association Studies (GWAS) Focusing on Patients with Mood or Anxiety Disorders and Co-Occurring Alcohol-Use Disorders. Diagnostics, 11(6), 1055. https://doi.org/10.3390/diagnostics11061055







