Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review
Abstract
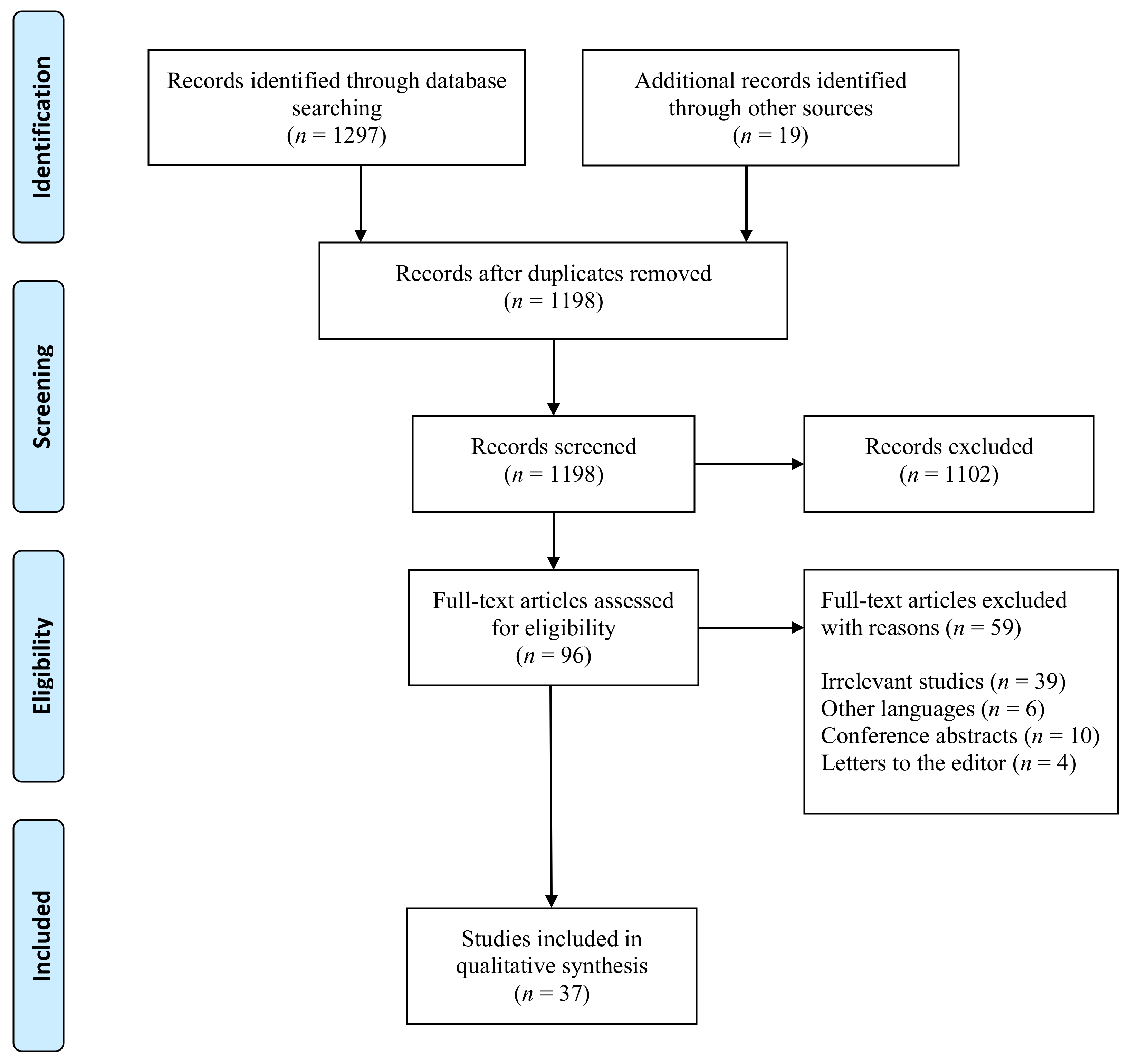
1. Introduction
2. Materials and Methods
3. Results
3.1. Artificial Intelligence Based Imaging for NAFLD Diagnosis
3.2. NAFLD Diagnosis and Staging Using Digital Pathology
3.3. NAFLD Diagnosis, Staging, and Risk Stratification Using Electronic Health Records
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sporea, I.; Popescu, A.; Dumitrașcu, D.; Brisc, C.; Nedelcu, L.; Trifan, A.; Gheorghe, L.; Fierbințeanu Braticevici, C. Nonalcoholic Fatty Liver Disease: Status Quo. J. Gastrointestin. Liver Dis. 2018, 27, 439–448. [Google Scholar] [PubMed]
- Sporea, I.; Sirli, R. Nonalcoholic Fatty Liver Disease and the Need for Action. J. Gastrointestin. Liver Dis. 2020, 29, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Cobbina, E.; Akhlaghi, F. Non-alcoholic fatty liver disease (NAFLD) - pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Bellentani, S. The epidemiology of non-alcoholic fatty liver disease. Liver Int. 2017, 37 (Suppl 1), 81–84. [Google Scholar] [CrossRef]
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62, S47–S64. [Google Scholar] [CrossRef]
- Calzadilla Bertot, L.; Adams, L.A. The Natural Course of Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2016, 17, 774. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar]
- Jones, L.D.; Golan, D.; Hanna, S.A.; Ramachandran, M. Artificial intelligence, machine learning and the evolution of healthcare: A bright future or cause for concern? Bone Joint Res. 2018, 7, 223–225. [Google Scholar] [CrossRef]
- Bhattad, P.B.; Jain, V. Artificial Intelligence in Modern Medicine—The Evolving Necessity of the Present and Role in Transforming the Future of Medical Care. Cureus 2020, 12, e8041. [Google Scholar] [CrossRef]
- Reyes, M.; Meier, R.; Pereira, S.; Silva, C.A.; Dahlweid, F.-M.; von Tengg-Kobligk, H.; Summers, R.M.; Wiest, R. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiol. Artif. Intell. 2020, 2, e190043. [Google Scholar] [CrossRef]
- Mihăilescu, D.M.; Gui, V.; Toma, C.I.; Popescu, A.; Sporea, I. Computer aided diagnosis method for steatosis rating in ultrasound images using random forests. Med. Ultrason. 2013, 15, 184–190. [Google Scholar] [CrossRef]
- Cao, W.; An, X.; Cong, L.; Lyu, C.; Zhou, Q.; Guo, R. Application of Deep Learning in Quantitative Analysis of 2-Dimensional Ultrasound Imaging of Nonalcoholic Fatty Liver Disease. J. Ultrasound Med. 2020, 39, 51–59. [Google Scholar] [CrossRef]
- Acharya, U.R.; Raghavendra, U.; Fujita, H.; Hagiwara, Y.; Koh, J.E.; Jen Hong, T.; Sudarshan, V.K.; Vijayananthan, A.; Yeong, C.H.; Gudigar, A.; et al. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. Comput. Biol. Med. 2016, 79, 250–258. [Google Scholar] [CrossRef]
- Biswas, M.; Kuppili, V.; Edla, D.R.; Suri, H.S.; Saba, L.; Marinhoe, R.T.; Sanches, J.M.; Suri, J.S. Symtosis: A liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput. Methods Programs Biomed. 2018, 155, 165–177. [Google Scholar] [CrossRef]
- Ribeiro, R.T.; Marinho, R.T.; Sanches, J.M. An ultrasound-based computer-aided diagnosis tool for steatosis detection. IEEE J. Biomed. Heal. Inform. 2014, 18, 1397–1403. [Google Scholar] [CrossRef]
- Kuppili, V.; Biswas, M.; Sreekumar, A.; Suri, H.S.; Saba, L.; Edla, D.R.; Marinho, R.T.; Sanches, J.M.; Suri, J.S. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J. Med. Syst. 2017, 41, 152. [Google Scholar] [CrossRef]
- Nagy, G.; Munteanu, M.; Gordan, M.; Chira, R.; Iancu, M.; Crisan, D.; Mircea, P.A. Computerized ultrasound image analysis for noninvasive evaluation of hepatic steatosis. Med. Ultrason. 2015, 17, 431–436. [Google Scholar]
- Subramanya, M.B.; Kumar, V.; Mukherjee, S.; Saini, M. A CAD system for B-mode fatty liver ultrasound images using texture features. J. Med. Eng. Technol. 2015, 39, 123–130. [Google Scholar] [CrossRef]
- Han, A.; Byra, M.; Heba, E.; Andre, M.P.; Erdman, J.W.; Loomba, R.; Sirlin, C.B.; O’Brien, W.D. Noninvasive Diagnosis of Nonalcoholic Fatty Liver Disease and Quantification of Liver Fat with Radiofrequency Ultrasound Data Using One-dimensional Convolutional Neural Networks. Radiology 2020, 295, 342–350. [Google Scholar] [CrossRef]
- Graffy, P.M.; Sandfort, V.; Summers, R.M.; Pickhardt, P.J. Automated Liver Fat Quantification at Nonenhanced Abdominal CT for Population-based Steatosis Assessment. Radiology 2019, 293, 334–342. [Google Scholar] [CrossRef]
- Jirapatnakul, A.; Reeves, A.P.; Lewis, S.; Chen, X.; Ma, T.; Yip, R.; Chin, X.; Liu, S.; Perumalswami, P.V.; Yankelevitz, D.F.; et al. Automated measurement of liver attenuation to identify moderate-to-severe hepatic steatosis from chest CT scans. Eur. J. Radiol. 2020, 122, 108723. [Google Scholar] [CrossRef]
- Huo, Y.; Terry, J.G.; Wang, J.; Nair, S.; Lasko, T.A.; Freedman, B.I.; Carr, J.J.; Landman, B.A. Fully automatic liver attenuation estimation combing CNN segmentation and morphological operations. Med. Phys. 2019, 46, 3508–3519. [Google Scholar] [CrossRef]
- De Rudder, M.; Bouzin, C.; Nachit, M.; Louvegny, H.; Vande Velde, G.; Julé, Y.; Leclercq, I.A. Automated computerized image analysis for the user-independent evaluation of disease severity in preclinical models of NAFLD/NASH. Lab. Investig. 2020, 100, 147–160. [Google Scholar] [CrossRef]
- Starke, A.; Haudum, A.; Weijers, G.; Herzog, K.; Wohlsein, P.; Beyerbach, M.; de Korte, C.L.; Thijssen, J.M.; Rehage, J. Noninvasive detection of hepatic lipidosis in dairy cows with calibrated ultrasonographic image analysis. J. Dairy Sci. 2010, 93, 2952–2965. [Google Scholar] [CrossRef]
- Acorda, J.A.; Yamada, H.; Ghamsari, S.M. Comparative evaluation of fatty infiltration of the liver in dairy cattle by using blood and serum analysis, ultrasonography, and digital analysis. Vet. Q. 1995, 17, 12–14. [Google Scholar] [CrossRef]
- Griffin, J.; Treanor, D. Digital pathology in clinical use: Where are we now and what is holding us back? Histopathology 2017, 70, 134–145. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Stathonikos, N.; van Varsseveld, N.C.; Vink, A.; van Dijk, M.R.; Nguyen, T.Q.; de Leng, W.W.J.; Lacle, M.M.; Goldschmeding, R.; Vreuls, C.P.H.; van Diest, P.J. Digital pathology in the time of corona. J. Clin. Pathol. 2020, 73, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Halicek, M.; Fabelo, H.; Callico, G.M.; Fei, B. Hyperspectral and multispectral imaging in digital and computational pathology: A systematic review [Invited]. Biomed. Opt. Express 2020, 11, 3195–3233. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, M.L.; Kolman, W.A.; Perry, B.; Prewitt, J.M. Computer analysis of cell images. Postgrad. Med. 1965, 38, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Forlano, R.; Mullish, B.H.; Giannakeas, N.; Maurice, J.B.; Angkathunyakul, N.; Lloyd, J.; Tzallas, A.T.; Tsipouras, M.; Yee, M.; Thursz, M.R.; et al. High-Throughput, Machine Learning-Based Quantification of Steatosis, Inflammation, Ballooning, and Fibrosis in Biopsies From Patients With Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2020, 18, 2081–2090.e9. [Google Scholar] [CrossRef]
- Munsterman, I.D.; van Erp, M.; Weijers, G.; Bronkhorst, C.; de Korte, C.L.; Drenth, J.P.H.; van der Laak, J.A.W.M.; Tjwa, E.T.T.L. A Novel Automatic Digital Algorithm that Accurately Quantifies Steatosis in NAFLD on Histopathological Whole-Slide Images. Cytom. B Clin. Cytom. 2019, 96, 521–528. [Google Scholar] [CrossRef]
- Segovia-Miranda, F.; Morales-Navarrete, H.; Kücken, M.; Moser, V.; Seifert, S.; Repnik, U.; Rost, F.; Brosch, M.; Hendricks, A.; Hinz, S.; et al. Three-dimensional spatially resolved geometrical and functional models of human liver tissue reveal new aspects of NAFLD progression. Nat. Med. 2019, 25, 1885–1893. [Google Scholar] [CrossRef]
- Vanderbeck, S.; Bockhorst, J.; Kleiner, D.; Komorowski, R.; Chalasani, N.; Gawrieh, S. Automatic quantification of lobular inflammation and hepatocyte ballooning in nonalcoholic fatty liver disease liver biopsies. Hum. Pathol. 2015, 46, 767–775. [Google Scholar] [CrossRef]
- Teramoto, T.; Shinohara, T.; Takiyama, A. Computer-aided classification of hepatocellular ballooning in liver biopsies from patients with NASH using persistent homology. Comput. Methods Programs Biomed. 2020, 195, 105614. [Google Scholar] [CrossRef]
- Gawrieh, S.; Sethunath, D.; Cummings, O.W.; Kleiner, D.E.; Vuppalanchi, R.; Chalasani, N.; Tuceryan, M. Automated quantification and architectural pattern detection of hepatic fibrosis in NAFLD. Ann. Diagn. Pathol. 2020, 47, 151518. [Google Scholar] [CrossRef]
- Vanderbeck, S.; Bockhorst, J.; Komorowski, R.; Kleiner, D.E.; Gawrieh, S. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum. Pathol. 2014, 45, 785–792. [Google Scholar] [CrossRef]
- Ramot, Y.; Zandani, G.; Madar, Z.; Deshmukh, S.; Nyska, A. Utilization of a Deep Learning Algorithm for Microscope-Based Fatty Vacuole Quantification in a Fatty Liver Model in Mice. Toxicol. Pathol. 2020, 48, 702–707. [Google Scholar] [CrossRef]
- Ge, F.; Lobdell, H.; Zhou, S.; Hu, C.; Berk, P.D. Digital analysis of hepatic sections in mice accurately quantitates triglycerides and selected properties of lipid droplets. Exp. Biol. Med. 2010, 235, 1282–1286. [Google Scholar] [CrossRef]
- Sethunath, D.; Morusu, S.; Tuceryan, M.; Cummings, O.W.; Zhang, H.; Yin, X.-M.; Vanderbeck, S.; Chalasani, N.; Gawrieh, S. Automated assessment of steatosis in murine fatty liver. PLoS One 2018, 13, e0197242. [Google Scholar] [CrossRef]
- Gunter, T.D.; Terry, N.P. The emergence of national electronic health record architectures in the United States and Australia: Models, costs, and questions. J. Med. Internet Res. 2005, 7, e3. [Google Scholar] [CrossRef]
- Van Vleck, T.T.; Chan, L.; Coca, S.G.; Craven, C.K.; Do, R.; Ellis, S.B.; Kannry, J.L.; Loos, R.J.F.; Bonis, P.A.; Cho, J.; et al. Augmented intelligence with natural language processing applied to electronic health records for identifying patients with non-alcoholic fatty liver disease at risk for disease progression. Int. J. Med. Inform. 2019, 129, 334–341. [Google Scholar] [CrossRef]
- Corey, K.E.; Kartoun, U.; Zheng, H.; Shaw, S.Y. Development and Validation of an Algorithm to Identify Nonalcoholic Fatty Liver Disease in the Electronic Medical Record. Dig. Dis. Sci. 2016, 61, 913–919. [Google Scholar] [CrossRef]
- Yip, T.C.-F.; Ma, A.J.; Wong, V.W.-S.; Tse, Y.-K.; Chan, H.L.-Y.; Yuen, P.-C.; Wong, G.L.-H. Laboratory parameter-based machine learning model for excluding non-alcoholic fatty liver disease (NAFLD) in the general population. Aliment. Pharmacol. Ther. 2017, 46, 447–456. [Google Scholar] [CrossRef]
- Perveen, S.; Shahbaz, M.; Keshavjee, K.; Guergachi, A. A Systematic Machine Learning Based Approach for the Diagnosis of Non-Alcoholic Fatty Liver Disease Risk and Progression. Sci. Rep. 2018, 8, 2112. [Google Scholar] [CrossRef]
- Katsiki, N.; Gastaldelli, A.; Mikhailidis, D.P. Predictive models with the use of omics and supervised machine learning to diagnose non-alcoholic fatty liver disease: A “non-invasive alternative” to liver biopsy? Metabolism 2019, 101, 154010. [Google Scholar] [CrossRef]
- Islam, M.M.; Wu, C.-C.; Poly, T.N.; Yang, H.-C.; Li, Y.-C.J. Applications of Machine Learning in Fatty Live Disease Prediction. Stud. Health Technol. Inform. 2018, 247, 166–170. [Google Scholar]
- Fialoke, S.; Malarstig, A.; Miller, M.R.; Dumitriu, A. Application of Machine Learning Methods to Predict Non-Alcoholic Steatohepatitis (NASH) in Non-Alcoholic Fatty Liver (NAFL) Patients. AMIA Annu. Symp. Proc. 2018, 2018, 430–439. [Google Scholar]
| First Author | Publication Year | Country | Study Design | Total Patients | Mean Age (Years) (Mean ± Sd) /(Range) | Diagnosis | Sex (% Male) | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Cao et al. [15] | 2019 | China | Cohort study | 240 | – | Nonalcoholic fatty liver disease (NAFLD) | 54.58% male | For the diagnosis of NAFLD, envelope signal and grayscale values are very important. Deep learning presented the best results in assessing the severity of NAFLD. |
| Acharya et al. [16] | 2016 | Malaysia | Retrospective cohort study | 150 | 22–79 years | Fatty liver disease | 50% | A new technique which uses a probabilistic neural network (PNN) classifier, that can automatically identify NAFLD with an accuracy of 97.33%, specificity of 100.00%, and sensitivity of 96.00%. |
| Biswas et al. [17] | 2017 | Portugal | Cohort study | 63 | – | Normal (n = 27) Fatty liver disease (n = 36) | – | Deep Learning-based Symtosis™ system is reliable and stable in comparison with support vector machine (SVM) and extreme learning machine (ELM) for the ultrasound categorization of hepatic tissue and risk stratification of normal and abnormal hepatic images containing hyper- and hypoechoic areas. |
| Ribeiro et al. [18] | 2014 | Portugal | Cohort study | 74 | – | Normal (n = 38) Hepatic steatosis (n = 36) | – | A new computer-aided diagnosis (CAD) system reported values of 93.33% for accuracy, 94.59% for sensitivity, and 92.11% for specificity, utilizing the Bayes classifier for steatosis detection and classification. |
| Kuppili et al. [19] | 2017 | Portugal | Cohort study | 63 | – | Normal (n = 27) Fatty liver disease (n = 36) | A tissue characterization system based on a class of Symtosis for risk stratification of ultrasonographic hepatic images demonstrates superior performance compared to SVM. | |
| Nagy et al. [20] | 2015 | Romania | Cohort study | 228 | 44 ± 11.38 | Mild hepatic steatosis (71.92%) Moderate-severe hepatic steatosis (28.08%) | 51.32% male | The development of an image analysis software that depicts three local intensity parameters: the coefficient of variation of luminance (CVL), the median luminance (ml), and the hepato-splenic attenuation index (HSAI) from regions of interest (ROI) in the ultrasound image and analyses their depth variation. The proposed computer analysis method was found to be of use for initial non-invasive assessment and grading of hepatic steatosis, with the added advantage of reduced complexity and accessibility for the computations. |
| Subramanya et al. [21] | 2014 | India | Cohort study | 53 | – | Normal liver subjects (n = 12) Fatty liver disease (n = 41)
| – | A computer-aided diagnosis (CAD) system for assessing hepatic steatosis grading, mainly mild, moderate, and severe fatty liver, as well as normal liver tissue was developed according to the visual interpretations of radiologists, that selected region of interests (ROIs) from within the liver and from the diaphragm region, in each image. The features of selected ROIs pertaining texture were three-way combined in order to obtain ratio features, inverse ratio features, and additive features. A DEFS (differential evolution feature selection) algorithm and a SVM have been used. The computer-aided diagnosis system had the potential of being of use to the radiologists in order to asses steatosis grades. |
| Mihailescu et al. [14] | 2013 | Romania | Cohort study | 120 | – | Normal liver subjects (n = 10) Fatty liver disease (n = 110)
| – | Random forests (RF) was superior to support vector machine (SVM) classifiers and proved the capability of this classifier to perform well without feature selection. On the other hand, the performance of the SVM classifier was low without feature selection, which improved greatly after feature selection. |
| Han et al. [22] | 2020 | USA | Single-center prospective study | 204 | NAFLD (52 years ± 14) Controls (46 years ± 21) | Non-alcoholic Fatty liver disease (n = 140) Controls (n = 64) | Non-alcoholic Fatty liver disease (58.57% male) Controls (65.63%) | Liver radiofrequency ultrasound data contains bountiful information regarding livers’ microstructure and its composition. Deep learning can utilize these informationx in order to to assess for the presence of nonalcoholic fatty liver disease (NAFLD). Deep learning algorithms utilizing radiofrequency ultrasound data are accurate both for diagnosing NAFLD and also for the quantification of hepatic fat fraction, given that other causes of steatosis were excluded. |
| Graffy et al. [23] | 2019 | USA | Retrospective cohort study | 9552 | 57.2 years ± 7.9 | No liver fat (n = 5584) Mild steatosis (n = 4948) Moderate steatosis (n = 1025) Severe steatosis (n = 112) | 44.36% male | The liver fat quantification tool, CT-based and fully automated, allows the population-based assessment of hepatic steatosis and NAFD, giving objective data that pairs well with data obtained by manual measurement. In this asymptomatic screening cohort, the prevalence of liver steatosis, of at least mild grade, was more than 50%. |
| Jirapatnakul et al. [24] | 2019 | USA | Cohort study | 333 | 65 years (IQR 54–69 years) 61 years (IQR 57–66 years) | Liver disease subjects (n = 24) Normal liver function subjects (n = 319) | 62.5% male 64.6% male | Automated measurements of liver attenuation in CT scans from patients with hepatic disease, can be utilized in identifying moderate to severe hepatic steatosis. The automated method outlines a set region within the liver, located below the right lung and utilizes statistical sampling techniques to elimitate all non-liver parenchyma. |
| Huo et al. [25] | 2019 | USA | Cohort study | 246 | – | NAFLD | – | An automatic ROI-based measurement (ALARM) method for estimating the liver attenuation was developed by combining DCNN and morphological operations, achieving an “excellent” accord with manual estimation of hepatic steatosis. The entire pipeline was implemented as a Docker container, enabling the software users to estimate the liver attenuation in five minutes per CT scan. |
| First Author | Publication Year | Country | Study Design | Total Patients | Mean Age (Years) (Mean ± Sd) /(Range) | Diagnosis | Sex (% Male) | Main Findings |
|---|---|---|---|---|---|---|---|---|
| De Rudder et al. [26] | 2019 | Belgium | Experimental study | 20 | – | NOD.B10 foz/foz and WT mice fed with high-fat diet C57B16/J mice fed with a fat-rich choline-deficient diet or fat-rich diet | 100% | The automated procedure was able to assess and measure the grade of macrovesicular steatosis, mixed inflammation, and pericellular fibrosis in steatohepatitis induced by CDAA. The procedure represents a promising quantitative technique with high-throughput, in order to rapidly evaluate the NAFLD activity in large preclinical studies, and for accurate survey of disease evolution. |
| Starke et al. [27] | 2010 | Germany | Experimental study | 151 | – | German Holstein dairy cows–normal | 0% | The CAUS methodology and software for digitally evaluating ultrasonographically obtained hepatic images could be accessible for noninvasive screening of hepatic steatosis in dairy herd health programs. Utilization of single parameter linear regression equation can possibly be optimal for practical applications. |
| Acorda et al. [28] | 2011 | Japan | Experimental study | 158 | – | Holstein-Friesian cows
| Fatty infiltration diagnosis was best performed using digital analysis, obtaining the highest sensitivity, specificity, accuracy, and PPV/NPV, followed by ultrasonography. |
| First Author | Publication Year | Country | Study Design | Total Patients | Mean Age (Years) (Mean ± Sd) /(Range) | Diagnosis | Sex (% Male) | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Griffin et al. [29] | 2017 | United Kingdom | Review | – | – | – | – | Combining whole slide imaging with other digital tools such as digital dictation, specimen trackin, and barcoding, can simplify the histopathology workflow, from specimen accession to report sign-out, enhancing the safety, quality, and efficiency of histopathology departments. |
| Bera et al. [30] | 2019 | USA | Review (Opinion) | – | – | – | – | The opportunity of digitalization whole-slide tissue images has contributed to the appearance of artificial intelligence and machine learning tools in digital pathology, enabling the search for subvisual morphometric phenotypes, hopefully improving patient management. |
| Stathonikos et al. [31] | 2020 | The Nederlands | Review | – | – | – | – | The 2020 COVID-19 crisis led to several implications affecting healthcare providers, including staffing shortages and the necessity to work from home. An asset digital diagnostic could allow pathologists, residents, molecular biologists, and pathology assistants to participate in the diagnostic process. |
| Ortega et al. [32] | 2020 | USA | Review | – | – | – | – | Using hyperspectral imaging (HSI) and multispectral imaging (MSI) technologies can add spatial information for creating computer-aided diagnostic tools for histological samples, both stained and unstained. HIS/MSI is associated with new possibilities in histological samples evaluation such as digital staining or mitigating the variability of digitized samples between different laboratories, compared to the traditional RGB analysis method. |
| Mendelsohn et al. [33] | 2016 | USA | Review | – | – | – | – | CYDAC (cytophotometric data converter) is a new modality, depending on computer analysis of cell images that can be applied to blood cell and chromosome discrimination. |
| This specific modality to pictorial data processing can possibly have utilizations in other scientific areas. | ||||||||
| Forlano et al. [34] | 2020 | United Kingdom | Retrospective cohort study | 246 | 51 (19–77) | Biopsy-proven NAFLD | 69% | An algorithm was developed through machine learning, by utilizing liver specimens obtained through biopsy from NAFLD patients, for quantifying the ammount of fat, inflammation, ballooning, and collagen. The algorithm proved predictability and sensitivity for the detection of modifications compared with traditional scores, in a cohort of paired specimens from liver biopsies. |
| Munsterman et al. [35] | 2019 | The Nederlands | Case-control study | 79 | – | Control subjects (n = 18) NAFLD subjects (n = 61) | – | Validation study for the development of a digital automated system for quantification of steatosis on whole-slide images (WSIs) of liver tissue. This algorithm can also be applied when appraising the degree of liver steatosis is warranted, such as clinical trials assessing the effectiveness of new therapeutic interventions in NAFLD. |
| Segovia-Miranda et al. [36] | 2019 | Germany | Cohort study | 25 | 68 (54–85) | Normal control (n = 6) | 33% | Computational simulations multiphoton imaging, and three-dimensional digital reconstructions were applied, achieving geometrical and functional spatially rendered models of human liver tissue from various non-alcoholic fatty liver disease (NAFLD) stages. The proposed models can define quantitative multiparametric signatures for cells and tissues, assessing disease progression and providing new patophysiologic insights into NAFLD. |
| 36.5 (29–68) | Healthy obese (n = 4) | 25% | ||||||
| 42 (34–51) | Steatosis (n = 8) | 63% | ||||||
| 51 (39–58) | Early NASH (n = 7) | 14% | ||||||
| Vanderbeck et al. [37] | 2015 | USA | Case-control study | 47 | – | Normal liver histology subjects (n = 20) NAFLD subjects (n = 27) of varying severity:
| – | Automatic quantification of cardinal NAFLD histologic lesions can be accessible and may possibly be used for further developing other automated methods for pathologists, in order to assess NAFLD biopsies in clinical practice and clinical trials. |
| Teramoto et al. [38] | 2020 | Japan | Cohort study | 80 | – | NAFLD subjects (n = 79) | – | A method linking topological data analysis with linear machine learning techniques was devised and tested for classifying liver tissue images, using Matteoni classification, into NAFLD subtypes. |
| Gawrieh et al. [39] | 2020 | USA | Cohort study | 18 | – | NAFLD (liver biopsies) | – | An integrated artificial intelligence-based automated tool was developed and tested to identify and evaluate hepatic fibrosis and its patterns in liver biopsies of NAFLD. |
| Vanderbeck et al. [40] | 2014 | USA | Case-control study | 47 | – | Normal liver histology (n = 20) NAFLD (n = 27) | – | An automatic classification of steatosis was proposed that included the basic features of NAFLD compared to other regions that appear as white in images of liver biopsies samples stained with hematoxylin and eosin (macrosteatosis, portal arteries, portal veins, central veins, sinusoids, and bile ducts). The precise identification of microscopic anatomical landmarks in liver samples, can ease critical ensuing tasks, (locating other histological anomalies) |
| First Author | Publication Year | Country | Study Design | Total Patients | Mean Age (Years) (Mean ± Sd) /(Range) | Diagnosis | Sex (% Male) | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Ramot et al. [41] | 2020 | Israel | Experimental study | 32 mice | – | Experiment with High-fat diet (HFD): 16 C57BL/6J male mice, divided in 6 groups: normal diet; HFD 10% (wt/wt) dietary broccoli; HFD10% (wt/wt) dietary broccoli stalks. Experiment with High cholesterol and cholate diet (HCD): 16 C57BL/6J male mice, divided in 4 groups: normal diet; diet high in fat high cholesterol (1%) and cholate (0.5%) (HCD; atherogenic diet); HCD 15% (wt/wt) dietary broccoli; HCD 15% (wt/wt) dietary broccoli stalks. | 100% | A deep learning AI algorithm was developed by utilizing glass slides of liver from mice models for nonalcoholic fatty liver disease, quantifying hepatic fatty vacuoles, while differentiating them from lumina of liver blood vessels and bile ducts. Utilizing deep learning algorithms for difficult assessments utilized in microscope-based pathology can advance outputs of workflows regarding toxicologic pathology. |
| Ge et al. [42] | 2010 | USA | Experimental study | 42 mice | Male C57BL/6J, ob/ob and db/db mice divided into 7 groups:
| – | A rapid and reproducible modality for histologic assessment of hepatic fat deposition in different models of hepatic steatosis in mice, based on quantitative digital analysis of Oil Red O (ORO) accumulation in fresh-frozen hepatic sections was developed. The process involved defining appropiate regions for analysis, followed by digital photographic imaging of these regions and subsequently by the digital determination of the portion of the identified area (Area Fraction) exhibiting ORO staining. | |
| Sethunath et al. [43] | 2018 | USA | Experimental study | 27 mice | – | Normal liver histology (n = 9) Mild steatosis (n = 10) Moderate steatosis (n = 4) Severe fatty liver (n = 4) | – | Accurate identification of macro- and microsteatosis in FLD in mice is critical in understanding the pathophysiology of the disease, and in the detection of potential hepatotoxic signatures which can be applied in drug development and quantifying the effects of different therapies. Automated classifiers were developed employing both image processing techniques and machine learning techniques, in order to study the correlation between automated quantification of macrosteatosis and semi-quantitative grades given by expert pathologists. The developed classifier proved high accuracy and sensitivity for indentifying macrosteatosis in fatty liver disease in mice. |
| First Author | Publication Year | Country | Study Design | Total Patients | Mean Age (Years) (Mean ± Sd) /(Range) | Diagnosis | Sex (% Male) | Main Findings |
|---|---|---|---|---|---|---|---|---|
| Van Vleck et al. [45] | 2019 | USA | Retrospective cohort study | 12,934 | 59.8 | NAFLD subjects (n = 2281) | 42% | The use of NLP (natural language processing) algorithms to appropiately evaluate unstructured data in EHR has been extensively documented. NLP-based approaches were more detailed in defining NAFLD within the EHR compared to ICD/text search-based methods. NLP algorithms promoted better analysis of knowledge flow between physician and allowed identifying certain breakdowns that conducted to key information misplacement, information that could have been used in slowing disease progression. |
| 59.5 | Control subjects (n = 10653) | 39% | ||||||
| Corey et al. [46] | 2016 | USA | Retrospective cohort study | 620 | 61.6 (12.7) | NAFLD subjects (n = 444) | 44% | The NAFLD classification algorithm designed within the electronic medical record (EMR) for establisment of longitudinal large-scale cohorts was better than ICD-9 billing data by itself. This method proved simple to establish and use through various institutions. |
| 58.7 (15.2) | Not NAFLD subjects (n = 176) | 46% | ||||||
| Yip et al. [47] | 2017 | Hong Kong | Retrospective cohort study | 922 | 50.7 ± 9.5 | NAFLD subjects (n = 264) | 54.2% | Six factors (ALT, HDL-C, triglyceride, HbA1c, white blood cell count, and hypertension) were included in a machine learning model based on laboratory parameter values, in order to identify NAFLD in the general population. The NAFLD ridge score was demonstrated to be a simple modality that can be compared to current NAFLD scores currently used in epidemiological studies to exclude patients with NAFLD |
| 47.0 ± 10.8 | Healthy subjects (n = 658) | 37.4% | ||||||
| Perveen et al. [48] | 2018 | Canada | Retrospective cohort study | 40,637 | 61.2 ± 14.2 | NAFLD subjects | 40% | A decision tree-based method was proposed for risk-assessment related to NAFLD development and progression in the Canadian population. The method used electronic medical eecords by analyzing the presence of risk factors for NAFLD. Using the proposed application linked to the everyday medical checkup is likely to aid physicians in performing more informed decisions about their patients’ management with NAFLD, while reducing healthcare spending. |
| Katsiki et al. [49] | 2019 | Greece, Italy, and United Kingdom | Editorial | – | – | – | – | Using lipidomic and metabolomic methods can contribute as diagnostic biomarkers because NAFLD is a metabolic disease. A pilot case–control study reported the benefits of utilizing non-invasive methods based on omics and supervised machine learning for diagnosing and treating NAFLD. These models may serve as a useful, non-invasive model, representing an attractive alternative to liver biopsy. |
| Islam et al. [50] | 2018 | Taiwan | Case-control study | 994 | 62.1 ± 12.55 | Fatty liver subjects (n = 593) | 46.37% | The study reports that machine learning models, especially logistic regression model are associated with an improved accuracy in predicting FLD, based on data found in electronic medical records and could be able to be an essential tool for clinical decision making. |
| 62.07 ± 13.52 | Non-fatty liver subjects (n = 401) | |||||||
| Fialoke et al. [51] | 2018 | USA | Retrospective cohort study | 108.139 | 57.60 (13.43) | NASH subjects (n = 17,359) | 41.22 | Longitudinal statistical properties of lab-based parameters (e.g., mean of all ALT values) were used to create supervised a machine learning model trained on NASH and healthy patients. The proposed model performed better than most of the non-invasive techniques currently in practice for diagnosing NASH. |
| 61.36 (18.34) | Healthy subjects (n = 17,590) | 40.17 | ||||||
| 57.30 (14.65) | Subjects at risk for NAFL (n = 73,190) | 39.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popa, S.L.; Ismaiel, A.; Cristina, P.; Cristina, M.; Chiarioni, G.; David, L.; Dumitrascu, D.L. Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review. Diagnostics 2021, 11, 1078. https://doi.org/10.3390/diagnostics11061078
Popa SL, Ismaiel A, Cristina P, Cristina M, Chiarioni G, David L, Dumitrascu DL. Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review. Diagnostics. 2021; 11(6):1078. https://doi.org/10.3390/diagnostics11061078
Chicago/Turabian StylePopa, Stefan L., Abdulrahman Ismaiel, Pop Cristina, Mogosan Cristina, Giuseppe Chiarioni, Liliana David, and Dan L. Dumitrascu. 2021. "Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review" Diagnostics 11, no. 6: 1078. https://doi.org/10.3390/diagnostics11061078
APA StylePopa, S. L., Ismaiel, A., Cristina, P., Cristina, M., Chiarioni, G., David, L., & Dumitrascu, D. L. (2021). Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review. Diagnostics, 11(6), 1078. https://doi.org/10.3390/diagnostics11061078








