Proteomic Exploration of Plasma Exosomes and Other Small Extracellular Vesicles in Pediatric Hodgkin Lymphoma: A Potential Source of Biomarkers for Relapse Occurrence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Research Ethics Statement
2.2. Patients and Plasma Samples
2.3. Study Design
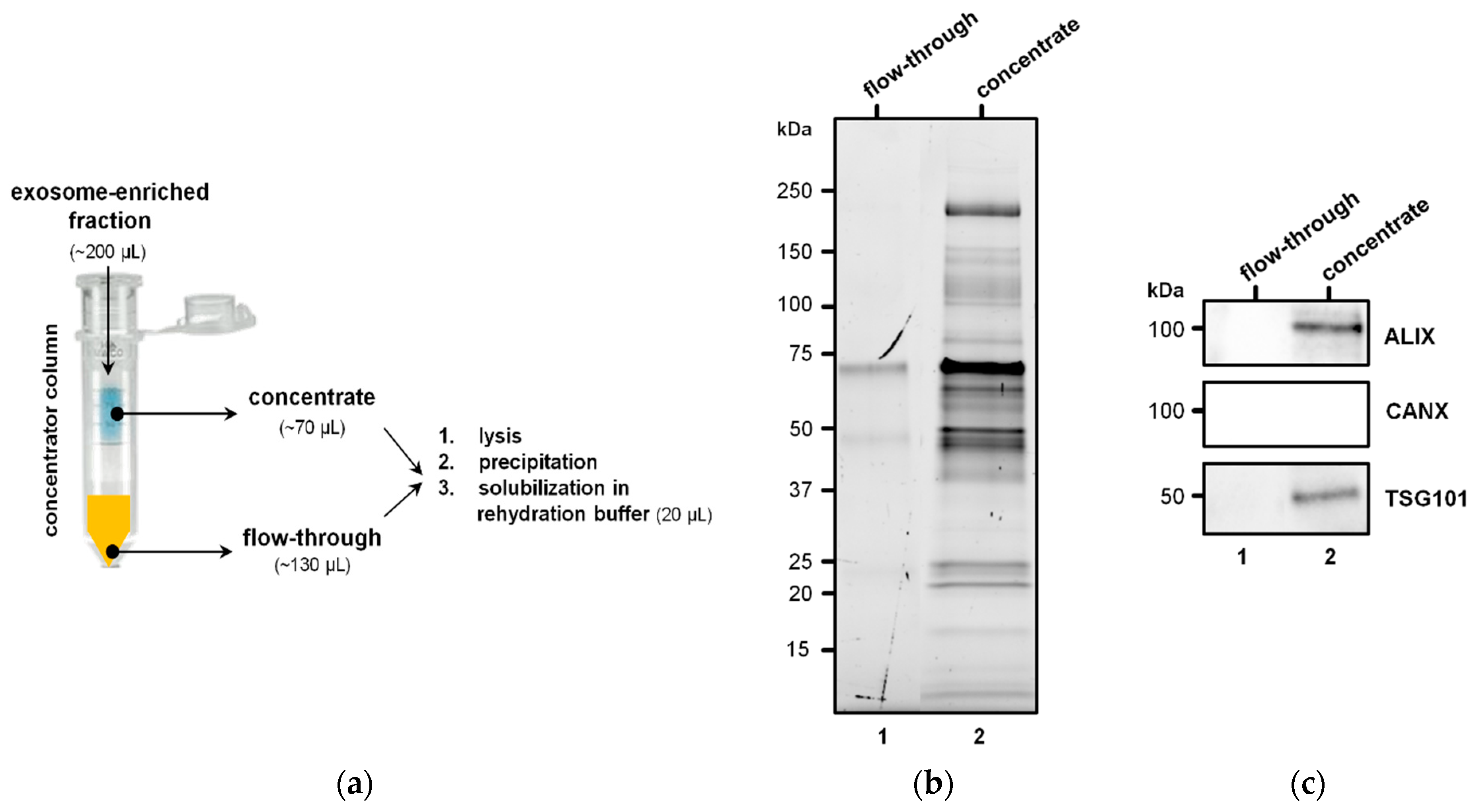
2.4. Isolation of EVs
2.5. Identification of Differentially Abundant Proteins
2.5.1. Protein Extraction
2.5.2. SDS-PAGE and Immunoblotting
2.5.3. Difference Gel Electrophoresis
2.5.4. Protein Identification by Mass Spectrometry
2.6. EV Protein Cargo Analysis in Non-Relapsed Pediatric HL
2.6.1. Protein Extraction
2.6.2. Protein Identification by Shotgun LC–MS/MS
2.6.3. Protein Functional Annotation
2.7. Interrogation of ExoCarta and Comparison with Previous Research
3. Results
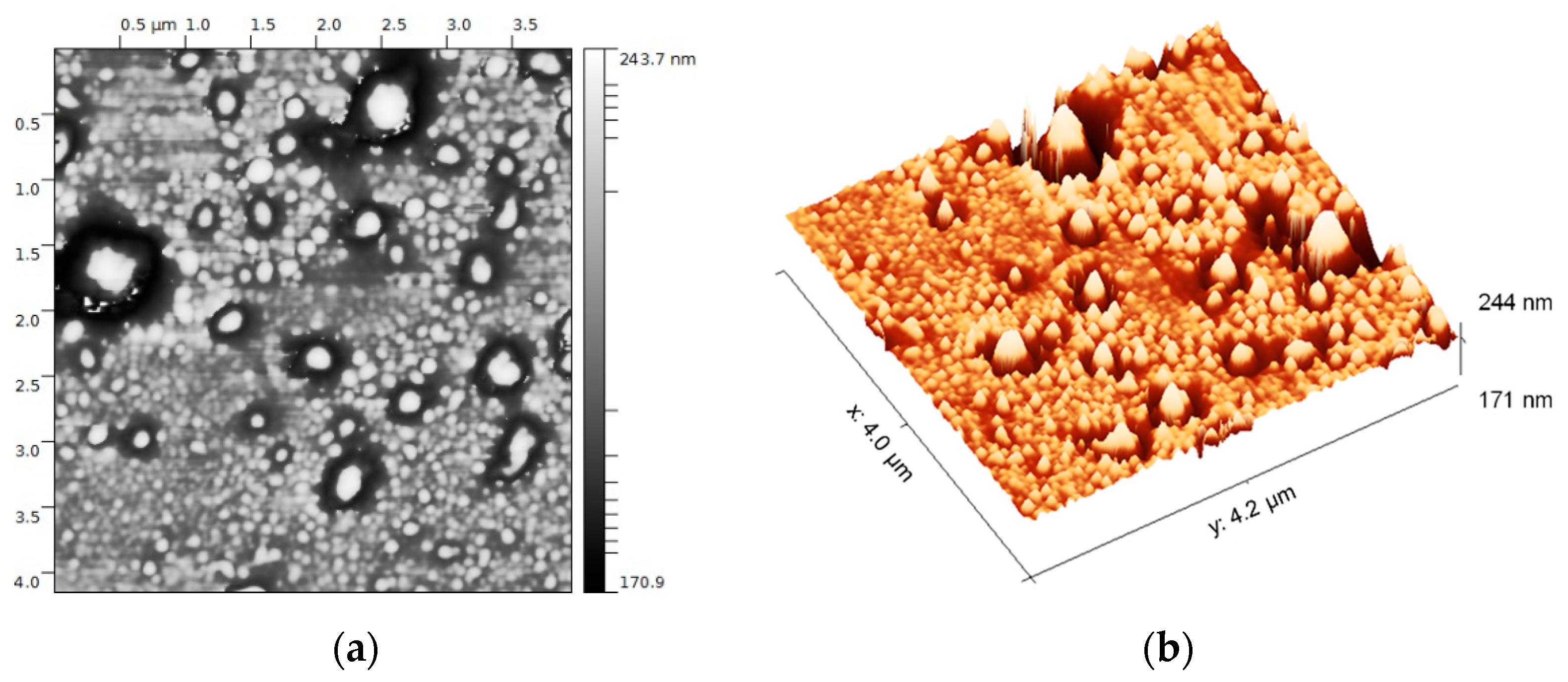
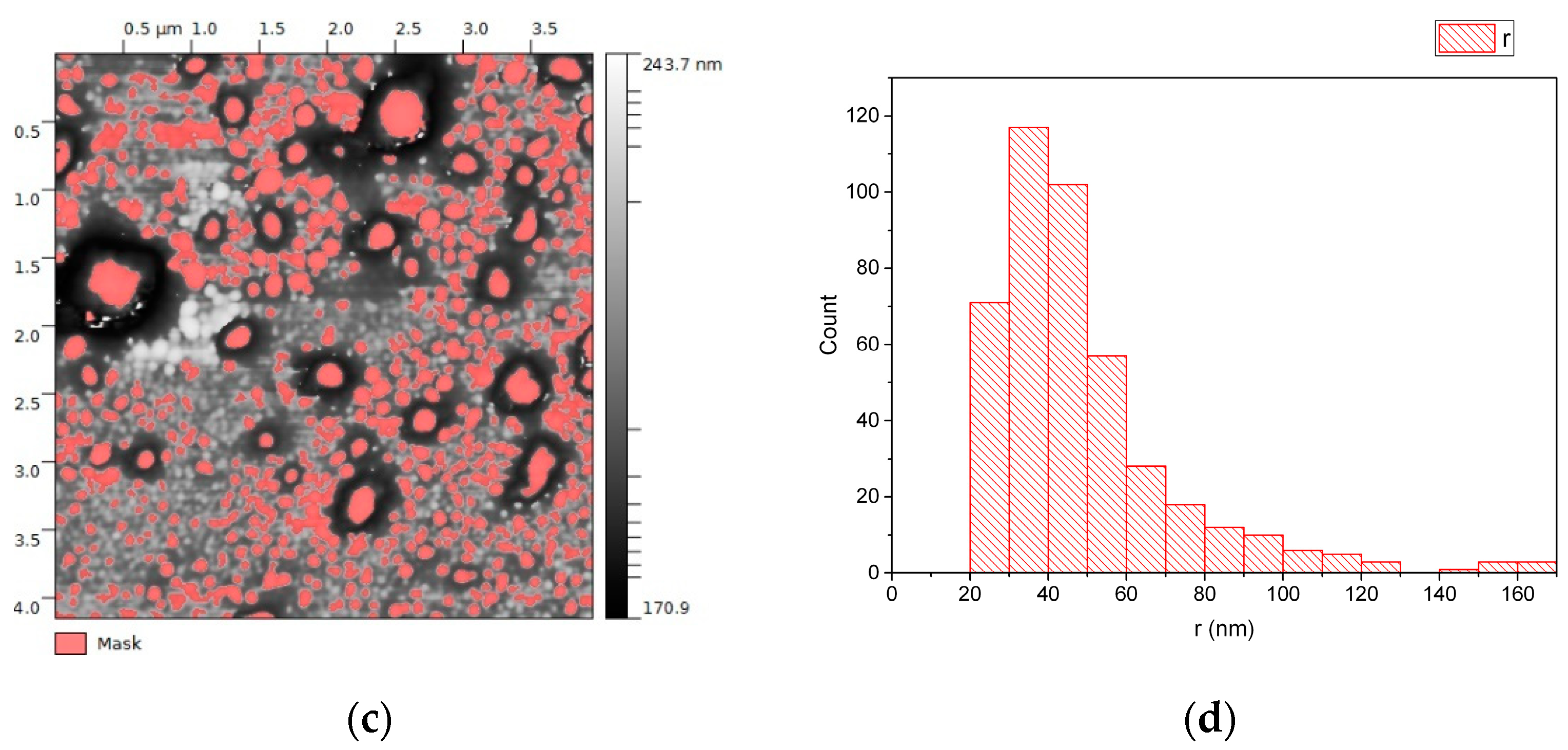
3.1. EVs from Plasma of Pediatric HL Patients
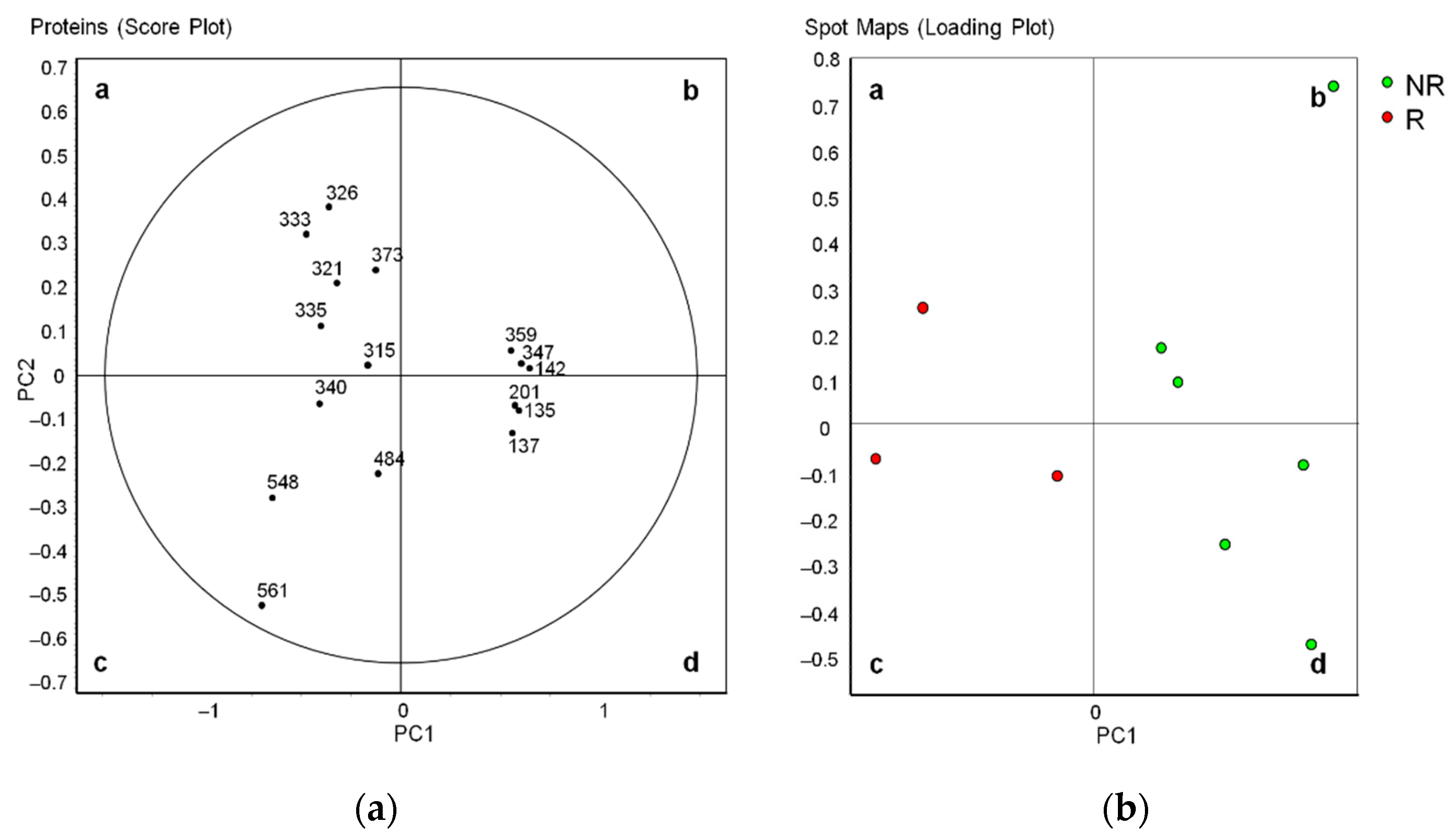
3.2. Differential Profiles of EV Proteins between Non-Relapsed and Relapsed HL
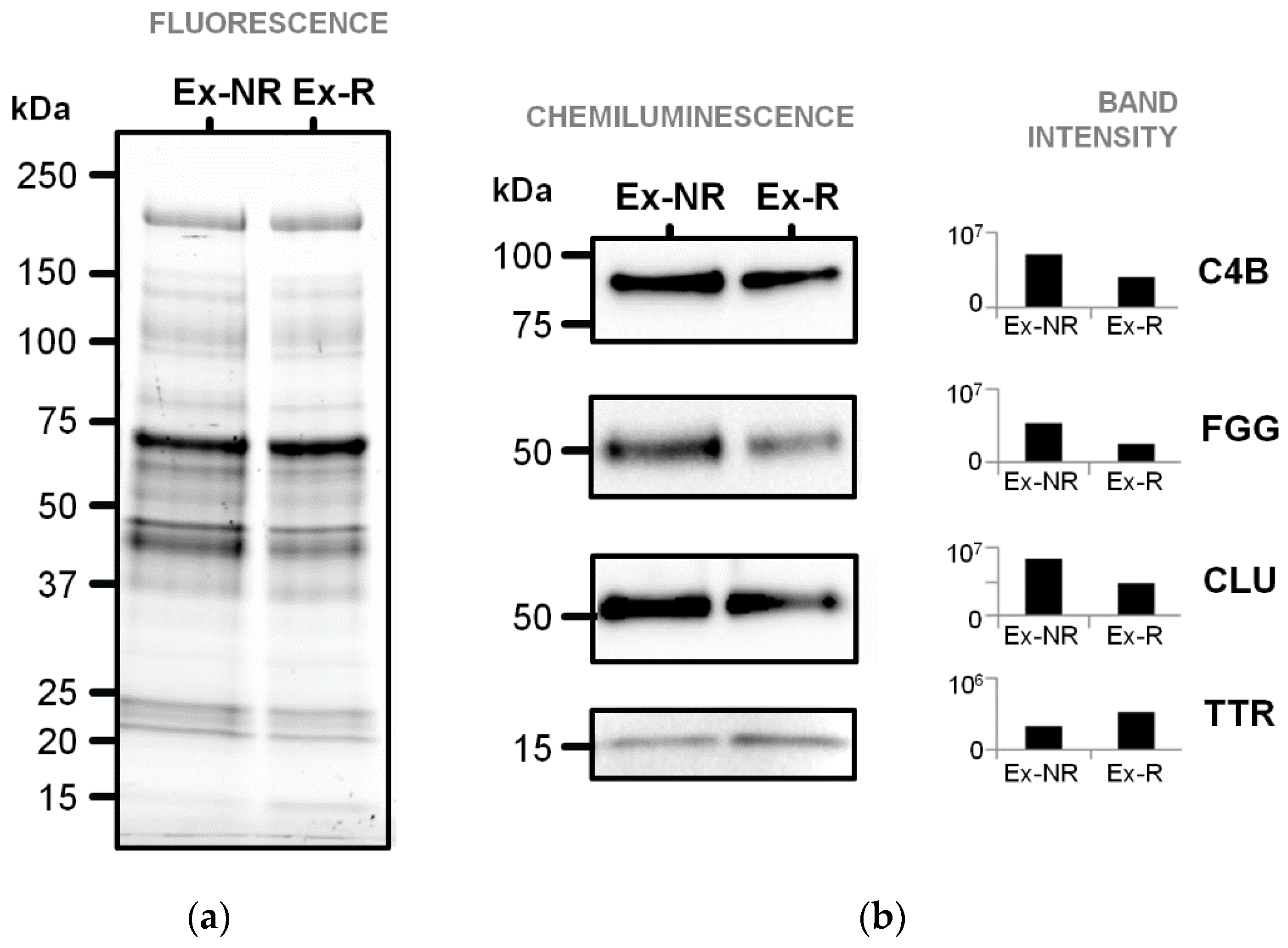
3.3. Immunoblotting Validation of Differentially Abundant Proteins
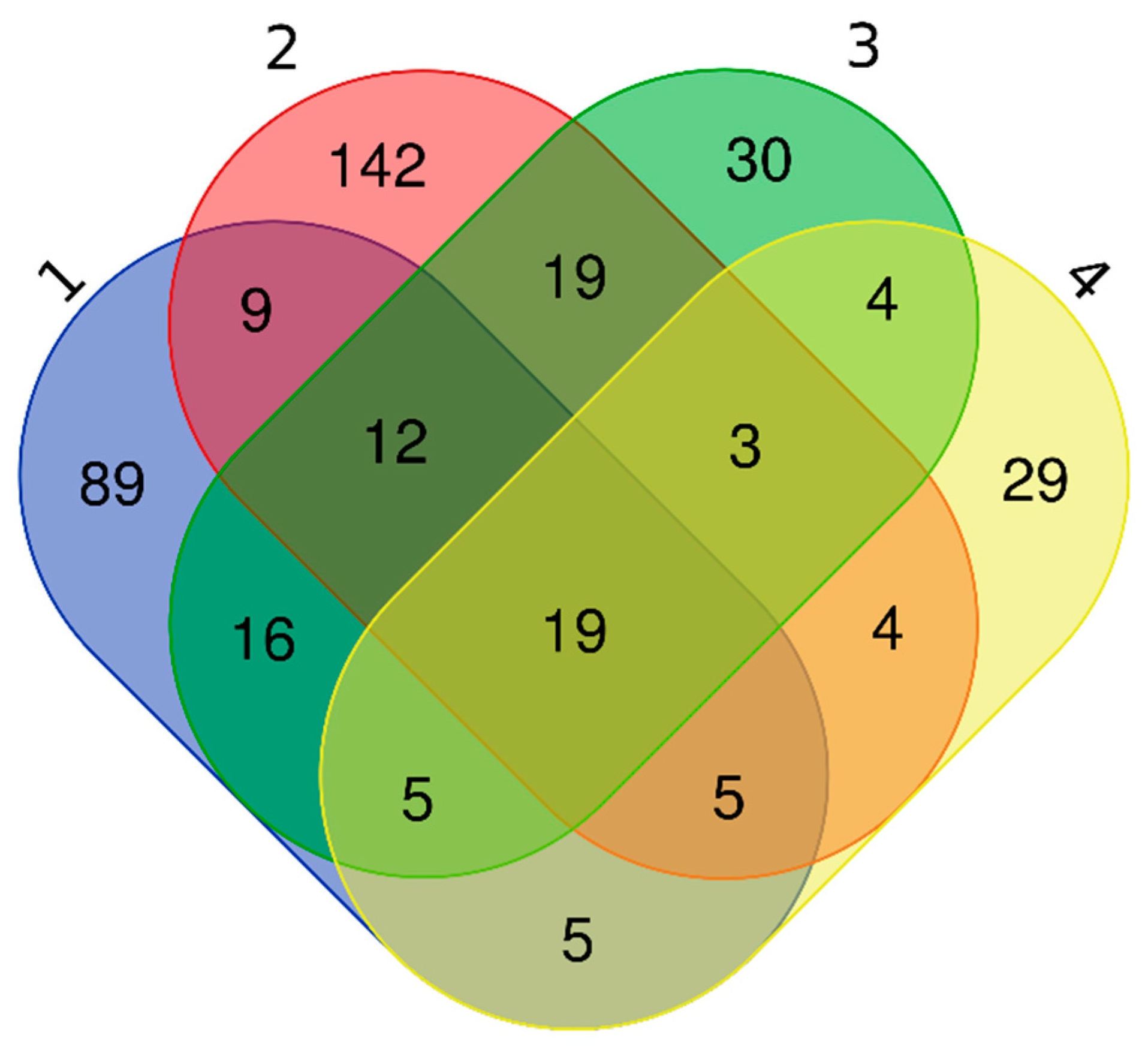
3.4. EV Protein Cargo in Non-Relapsed Pediatric HL
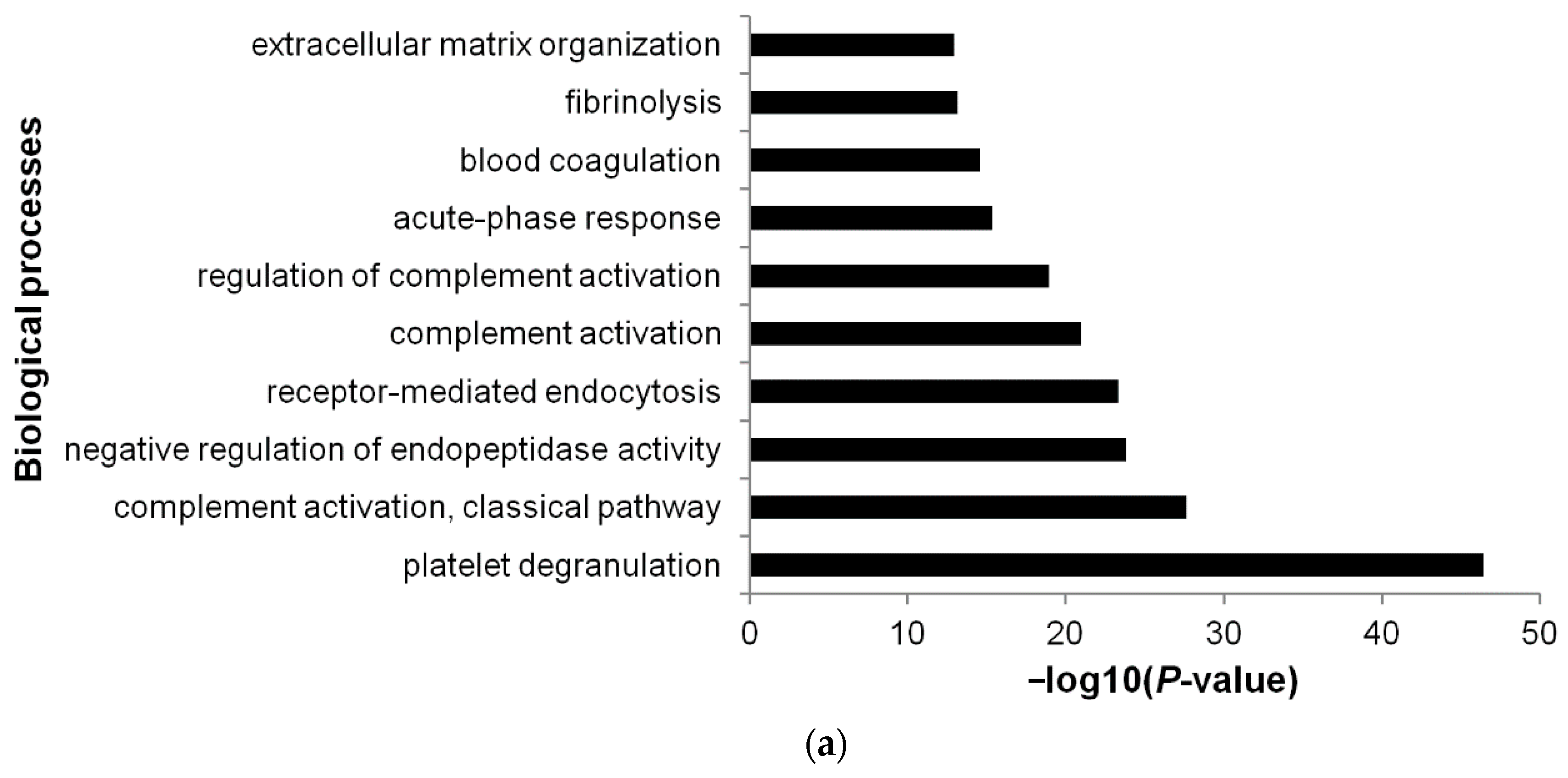
3.5. Functional Annotations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaatsch, P. Epidemiology of childhood cancer. Cancer Treat. Rev. 2010, 36, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Altekruse, S.F.; Adamson, P.C.; Reaman, G.H.; Seibel, N.L. Declining childhood and adolescent cancer mortality. Cancer 2014, 120, 2497–2506. [Google Scholar] [CrossRef]
- Daw, S.; Hasenclever, D.; Mascarin, M.; Fernández-Teijeiro, A.; Balwierz, W.; Beishuizen, A.; Burnelli, R.; Cepelova, M.; Claviez, A.; Dieckmann, K.; et al. Risk and response adapted treatment guidelines for managing first relapsed and refractory classical hodgkin lymphoma in children and young people. recommendations from the euronet pediatric hodgkin lymphoma group. HemaSphere 2020, 4. [Google Scholar] [CrossRef]
- Schwartz, C.L.; Constine, L.S.; Villaluna, D.; London, W.B.; Hutchison, R.E.; Sposto, R.; Lipshultz, S.E.; Turner, C.S.; deAlarcon, P.A.; Chauvenet, A. A risk-adapted, response-based approach using ABVE-PC for children and adolescents with intermediate- and high-risk odgkin lymphoma: The results of P9425. Blood 2009, 114, 2051–2059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repetto, O.; De Re, V.; Mussolin, L.; Tedeschi, M.; Elia, C.; Bianchi, M.; Buffardi, S.; Sala, A.; Burnelli, R.; Mascarin, M. Proteomic profiles and biological processes of relapsed vs. non-relapsed pediatric hodgkin lymphoma. Int. J. Mol. Sci. 2020, 21, 2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repetto, O.; Mussolin, L.; Elia, C.; Martina, L.; Bianchi, M.; Buffardi, S.; Sala, A.; Burnelli, R.; Mascarin, M.; De Re, V. Proteomic identification of plasma biomarkers in children and adolescents with recurrent Hodgkin lymphoma. J. Cancer 2018, 9, 4650–4658. [Google Scholar] [CrossRef]
- Shafat, I.; Barak, A.B.; Postovsky, S.; Elhasid, R.; Ilan, N.; Vlodavsky, I.; Arush, M.W.B. Heparanase levels are elevated in the plasma of pediatric cancer patients and correlate with response to anticancer treatment. Neoplasia 2007, 9, 909–916. [Google Scholar] [CrossRef] [Green Version]
- Pui, C.H.; Hudson, M.; Luo, X.; Wilimas, J.; Evans, W.; Crist, W.M. Serum interleukin-2 receptor levels in hodgkin disease and other solid tumors of childhood. Leukemia 1993, 7, 1242–1244. [Google Scholar]
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol. Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef]
- Garcia-Romero, N.; Esteban-Rubio, S.; Rackov, G.; Carrión-Navarro, J.; Belda-Iniesta, C.; Ayuso-Sacido, A. Extracellular vesicles compartment in liquid biopsies: Clinical application. Mol. Aspects Med. 2018, 60, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hristov, M.; Erl, W.; Linder, S.; Weber, P.C. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood 2004, 104, 2761–2766. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.S.; Madala, S.K.; Trinath, J.; Reddy, G.B. Extracellular small heat shock proteins: Exosomal biogenesis and function. Cell Stress Chaperones 2018, 23, 441–454. [Google Scholar] [CrossRef]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Segura, E.; Amigorena, S.; Théry, C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood Cells. Mol. Dis. 2005, 35, 89–93. [Google Scholar] [CrossRef]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of MRNAs and MicroRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Denzer, K.; van Eijk, M.; Kleijmeer, M.J.; Jakobson, E.; de Groot, C.; Geuze, H.J. Follicular dendritic cells carry MHC class ii-expressing microvesicles at their surface. J. Immunol. 2000, 165, 1259–1265. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Lorenzo, M.J.; Anel, A.; Gamen, S.; Monlen, I.; Lasierra, P.; Larrad, L.; Piñeiro, A.; Alava, M.A.; Naval, J. Activated human t cells release bioactive FAS ligand and APO2 ligand in microvesicles. J. Immunol. 1999, 163, 1274–1281. [Google Scholar]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heijnen, H.F.G.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Federici, C.; Shahaj, E.; Cecchetti, S.; Camerini, S.; Casella, M.; Iessi, E.; Camisaschi, C.; Paolino, G.; Calvieri, S.; Ferro, S.; et al. Natural-killer-derived extracellular vesicles: Immune sensors and interactors. Front. Immunol. 2020, 11, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, S.C.; Nadler, E.P.; Pillai, D.K.; Hubal, M.J.; Wang, Z.; Wang, J.M.; Gordish-Dressman, H.; Koeck, E.; Sevilla, S.; Wiles, A.A.; et al. Adipocyte-derived exosomal MiRNAs: A novel mechanism for obesity-related disease. Pediatr. Res. 2015, 77, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.; Wang, H.; Li, B.; Qin, Q.; Qi, L.; Nagarkatti, M.; Nagarkatti, P.; Janicki, J.S.; Wang, X.L.; Cui, T. A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J. Mol. Cell. Cardiol. 2015, 89, 268–279. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367. [Google Scholar] [CrossRef]
- Inal, J.M.; Kosgodage, U.; Azam, S.; Stratton, D.; Antwi-Baffour, S.; Lange, S. Blood/plasma secretome and microvesicles. Biochim. Biophys. Acta 2013, 1834, 2317–2325. [Google Scholar] [CrossRef]
- Yuana, Y.; Sturk, A.; Nieuwland, R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013, 27, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.E.; Sanderson, R.D. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xu, M.; Lu, J.; Mao, L.; Wang, S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol. Cancer 2019, 18, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with Anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Rosenow, M.; Xiao, N.; Spetzler, D. Profiling plasma extracellular vesicle by pluronic block-copolymer based enrichment method unveils features associated with breast cancer aggression, metastasis and invasion. J. Extracell. Vesicles 2018, 7, 1458574. [Google Scholar] [CrossRef] [Green Version]
- Wen, S.W.; Sceneay, J.; Lima, L.G.; Wong, C.S.F.; Becker, M.; Krumeich, S.; Lobb, R.J.; Castillo, V.; Wong, K.N.; Ellis, S.; et al. The biodistribution and immune suppressive effects of breast cancer-derived exosomes. Cancer Res. 2016, 76, 6816–6827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Eijndhoven, M.A.; Zijlstra, J.M.; Groenewegen, N.J.; Drees, E.E.; van Niele, S.; Baglio, S.R.; Koppers-Lalic, D.; van der Voorn, H.; Libregts, S.F.; Wauben, M.H.; et al. Plasma vesicle MiRNAs for therapy response monitoring in hodgkin lymphoma patients. JCI Insight 2016, 1, e89631. [Google Scholar] [CrossRef]
- Hernández-Walias, F.J.; Vázquez, E.; Pacheco, Y.; Rodríguez-Fernández, J.M.; Pérez-Elías, M.J.; Dronda, F.; Casado, J.L.; Moreno, A.; Hermida, J.M.; Quereda, C.; et al. Risk, diagnostic and predictor factors for classical hodgkin lymphoma in HIV-1-infected individuals: Role of plasma exosome-derived MiR-20a and MiR-21. J. Clin. Med. 2020, 9, 760. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, D.P.; Dittmar, G. Peptide array-based interactomics. Anal. Bioanal. Chem. 2021. [Google Scholar] [CrossRef]
- Vinaiphat, A.; Low, J.K.; Yeoh, K.W.; Chng, W.J.; Sze, S.K. Application of advanced mass spectrometry-based proteomics to study hypoxia driven cancer progression. Front. Oncol. 2021, 11, 559822. [Google Scholar] [CrossRef]
- Vitorino, R.; Guedes, S.; Costa, J.P.D.; Kašička, V. Microfluidics for peptidomics, proteomics, and cell analysis. Nanomaterials 2021, 11, 1118. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.-A.; Park, J.-M.; Lee, H. Review of three-dimensional liquid chromatography platforms for bottom-up proteomics. Int. J. Mol. Sci. 2020, 21, 1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadlmann, J.; Hudecz, O.; Krššáková, G.; Ctortecka, C.; Van Raemdonck, G.; Op De Beeck, J.; Desmet, G.; Penninger, J.M.; Jacobs, P.; Mechtler, K. Improved sensitivity in low-input proteomics using micropillar array-based chromatography. Anal. Chem. 2019, 91, 14203–14207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexovič, M.; Sabo, J.; Longuespée, R. Microproteomic sample preparation. Proteomics 2021, 21, e2000318. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.T. Single-cell proteomics: Progress and prospects. Mol. Cell. Proteomics 2020, 19, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Xue, V.W.; Yang, C.; Wong, S.C.C.; Cho, W.C.S. Proteomic profiling in extracellular vesicles for cancer detection and monitoring. Proteomics 2021, e2000094. [Google Scholar] [CrossRef]
- Bandu, R.; Oh, J.W.; Kim, K.P. Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kuang, M.; Tao, X.; Peng, Y.; Zhang, W.; Pan, Y.; Cheng, L.; Yuan, C.; Zhao, Y.; Mao, H.; Zhuge, L.; et al. Proteomic analysis of plasma exosomes to differentiate malignant from benign pulmonary nodules. Clin. Proteom. 2019, 16. [Google Scholar] [CrossRef]
- Chen, I.-H.; Xue, L.; Hsu, C.-C.; Paez, J.S.P.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.-K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ou, X.; Wu, X. Proteomics profiling of plasma exosomes in epithelial ovarian cancer: A potential role in the coagulation cascade, diagnosis and prognosis. Int. J. Oncol. 2019, 54, 1719–1733. [Google Scholar] [CrossRef] [Green Version]
- Farruggia, P.; Puccio, G.; Sala, A.; Todesco, A.; Buffardi, S.; Garaventa, A.; Bottigliero, G.; Bianchi, M.; Zecca, M.; Locatelli, F.; et al. The prognostic value of biological markers in paediatric hodgkin lymphoma. Eur. J. Cancer 2016, 52, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood 2011, 117, 5019–5032. [Google Scholar] [CrossRef] [Green Version]
- Carbone, P.P.; Kaplan, H.S.; Musshoff, K.; Smithers, D.W.; Tubiana, M. Report of the committee on hodgkin’s disease staging classification. Cancer Res. 1971, 31, 1860–1861. [Google Scholar] [PubMed]
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS ONE 2015, 10, e0136133. [Google Scholar] [CrossRef] [Green Version]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the international society for extracellular vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [Green Version]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.-S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef]
- Bastos-Amador, P.; Royo, F.; Gonzalez, E.; Conde-Vancells, J.; Palomo-Diez, L.; Borras, F.E.; Falcon-Perez, J.M. Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J. Proteom. 2012, 75, 3574–3584. [Google Scholar] [CrossRef]
- Looze, C.; Yui, D.; Leung, L.; Ingham, M.; Kaler, M.; Yao, X.; Wu, W.W.; Shen, R.-F.; Daniels, M.P.; Levine, S.J. Proteomic profiling of human plasma exosomes identifies PPARγ as an exosome-associated protein. Biochem. Biophys. Res. Commun. 2009, 378, 433–438. [Google Scholar] [CrossRef] [Green Version]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Jonas, A. Lipoprotein structure. In Biochemistry of Lipids, Lipoproteins and Membranes, 4th ed.; Vance, D.E., Vance, J.E., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2002; Volume 36, pp. 483–504. [Google Scholar]
- Rensen, P.C.; de Vrueh, R.L.; Kuiper, J.; Bijsterbosch, M.K.; Biessen, E.A.; van Berkel, T.J. Recombinant lipoproteins: Lipoprotein-like lipid particles for drug targeting. Adv. Drug Deliv. Rev. 2001, 47, 251–276. [Google Scholar] [CrossRef]
- Kitamura, Y.; Kojima, M.; Kurosawa, T.; Sasaki, R.; Ichihara, S.; Hiraku, Y.; Tomimoto, H.; Murata, M.; Oikawa, S. Proteomic profiling of exosomal proteins for blood-based biomarkers in Parkinson’s disease. Neuroscience 2018, 392, 121–128. [Google Scholar] [CrossRef]
- Tsuno, H.; Arito, M.; Suematsu, N.; Sato, T.; Hashimoto, A.; Matsui, T.; Omoteyama, K.; Sato, M.; Okamoto, K.; Tohma, S.; et al. A proteomic analysis of serum-derived exosomes in rheumatoid arthritis. BMC Rheumatol. 2018, 2, 35. [Google Scholar] [CrossRef]
- Richardson, S.J. Evolutionary changes to transthyretin: Evolution of transthyretin biosynthesis. FEBS J. 2009, 276, 5342–5356. [Google Scholar] [CrossRef]
- Weisel, J.W. Fibrinogen and fibrin. Adv. Protein Chem. 2005, 70, 247–299. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, S.O.; Simpson-Haidaris, P.J. Regulated de novo biosynthesis of fibrinogen in extrahepatic epithelial cells in response to inflammation. Thromb. Haemost. 2004, 92, 234–243. [Google Scholar] [CrossRef]
- Ahmed, F.E. Utility of mass spectrometry for proteome Ana Lysis: Part I. conceptual and experimental approaches. Expert Rev. Proteom. 2008, 5, 841–864. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Jadhav, A.B.; Hassan, A.; Meng, Q.H. Acute phase reactants as novel predictors of cardiovascular disease. ISRN Inflamm. 2012, 2012, 953461. [Google Scholar] [CrossRef] [Green Version]
- Farrell, D.H. Pathophysiologic roles of the fibrinogen gamma chain. Curr. Opin. Hematol. 2004, 11, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Mosesson, M.W. Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 2005, 3, 1894–1904. [Google Scholar] [CrossRef]
- Karaosmanoğlu, O.; Banerjee, S.; Sivas, H. Identification of biomarkers associated with partial epithelial to mesenchymal transition in the secretome of slug over-expressing hepatocellular carcinoma cells. Cell. Oncol. 2018, 41, 439–453. [Google Scholar] [CrossRef]
- Repetto, O.; De Re, V. Coagulation and fibrinolysis in gastric cancer. Ann. N. Y. Acad. Sci. 2017, 1404, 27–48. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, Z.; Zeng, G.; Xie, H.; Liu, J.; Liu, N.; Wang, G. Clinical significance of urinary plasminogen and fibrinogen gamma chain as novel potential diagnostic markers for non-small-cell lung cancer. Clin. Chim. Acta Int. J. Clin. Chem. 2020, 502, 55–65. [Google Scholar] [CrossRef]
- Araújo, J.E.; López-Fernández, H.; Diniz, M.S.; Baltazar, P.M.; Pinheiro, L.C.; da Silva, F.C.; Carrascal, M.; Videira, P.; Santos, H.M.; Capelo, J.L. Dithiothreitol-based protein equalization technology to unravel biomarkers for bladder cancer. Talanta 2018, 180, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Gião, T.; Saavedra, J.; Cotrina, E.; Quintana, J.; Llop, J.; Arsequell, G.; Cardoso, I. Undiscovered roles for transthyretin: From a transporter protein to a new therapeutic target for Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, H.; Su, X.; Jin, Y.; Zhang, C.; Wang, L.; Yang, L. Transthyretin regulated by Linc00657/MiR-205-5p promoted cholesterol metabolism by inducing SREBP2-HMGCR and inhibiting LXRα-CYP7A1. Arch. Med. Res. 2020, 51, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Khan, S.; Rahman, S.; Singh, L.R. The extracellular protein, transthyretin is an oxidative stress biomarker. Front. Physiol. 2019, 10, 5. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Q.; Li, T.; Liao, Q.; Zhao, Y. Role of the complement system in the tumor microenvironment. Cancer Cell Int. 2019, 19, 300. [Google Scholar] [CrossRef] [PubMed]
- Vélez, P.; Parguiña, A.F.; Ocaranza-Sánchez, R.; Grigorian-Shamagian, L.; Rosa, I.; Alonso-Orgaz, S.; de la Cuesta, F.; Guitián, E.; Moreu, J.; Barderas, M.G.; et al. Identification of a circulating microvesicle protein network involved in st-elevation myocardial infarction. Thromb. Haemost. 2014, 112, 716–726. [Google Scholar] [CrossRef]
- Kuravi, S.J.; Harrison, P.; Rainger, G.E.; Nash, G.B. Ability of platelet-derived extracellular vesicles to promote neutrophil-endothelial cell interactions. Inflammation 2019, 42, 290–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef] [PubMed]
- Heutinck, K.M.; ten Berge, I.J.M.; Hack, C.E.; Hamann, J.; Rowshani, A.T. Serine proteases of the human immune system in health and disease. Mol. Immunol. 2010, 47, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Di Cera, E. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef]
- Das, A.; Mohan, V.; Krishnaswamy, V.R.; Solomonov, I.; Sagi, I. Exosomes as a storehouse of tissue remodeling proteases and mediators of cancer progression. Cancer Metastasis Rev. 2019, 38, 455–468. [Google Scholar] [CrossRef]
- Karasu, E.; Eisenhardt, S.U.; Harant, J.; Huber-Lang, M. Extracellular vesicles: Packages sent with complement. Front. Immunol. 2018, 9, 721. [Google Scholar] [CrossRef] [PubMed]
- Hiltbrunner, S.; Larssen, P.; Eldh, M.; Martinez-Bravo, M.-J.; Wagner, A.K.; Karlsson, M.C.I.; Gabrielsson, S. Exosomal cancer immunotherapy is independent of mhc molecules on exosomes. Oncotarget 2016, 7, 38707–38717. [Google Scholar] [CrossRef] [Green Version]
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Dillon, S.T.; Reeves, R.K.; Manickam, C.; Morgello, S.; Kirk, G.D.; Mehta, S.H.; Gabuzda, D. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Sci. Rep. 2018, 8, 7227. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B Lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Li, Q.; Wang, H.; Peng, H.; Huyan, T.; Cacalano, N.A. Exosomes: Versatile nano mediators of immune regulation. Cancers 2019, 11, 1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groot Kormelink, T.; Mol, S.; de Jong, E.C.; Wauben, M.H.M. The role of extracellular vesicles when innate meets adaptive. Semin. Immunopathol. 2018, 40, 439–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robbins, P.D.; Dorronsoro, A.; Booker, C.N. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J. Clin. Investig. 2016, 126, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enblad, G.; Molin, D.; Glimelius, I.; Fischer, M.; Nilsson, G. The potential role of innate immunity in the pathogenesis of Hodgkin’s lymphoma. Hematol. Oncol. Clin. N. Am. 2007, 21, 805–823. [Google Scholar] [CrossRef] [PubMed]





| Explorative Set (Part I: 2D-DIGE) | Validation Set (Part I: Immunoblotting; Part II: Shotgun LC–MS/MS) | |||
|---|---|---|---|---|
| Characteristic | Non-Relapsed HL (n = 6) | Relapsed HL (n = 3) | Non-Relapsed HL (n = 3) | Relapsed HL (n = 3) |
| Gender | ||||
| Male | 4 | 3 | 1 | 6 |
| Female | 2 | 0 | 2 | 0 |
| Age at diagnosis, years, mean (SD) | 14 (2) | 14 (3) | 15 (1) | 17 (1) |
| Stage, n a | ||||
| II | 3 | 2 | 3 | 0 |
| IV | 3 | 1 | 0 | 3 |
| Systemic symptoms, n | 3 | 2 | 1 | 2 |
| LH2004 therapeutic group | ||||
| 1 | 1 | 0 | 0 | 0 |
| 2 | 2 | 0 | 3 | 0 |
| 3 | 3 | 3 | 0 | 3 |
| Spot No. a | MW (pI) | Database | Accession | Gene | Protein | Score | Seq. cov. % b | Fold Change c | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| More abundant in non-relapsed HL (n = 6) | |||||||||
| 142 | 106,853 (6.40) | SwissProt | ITIH2_HUMAN | ITIH2 | Inter-α-trypsin inhibitor heavy chain H2 ** | 472 | 19 | −2.0 | 0.033 |
| 135 | 194,170 (6.89) | SwissProt | CO4B_HUMAN | C4B | Complement C4-B * | 351 | 6 | −2.0 | 0.018 |
| 137 | 189,125 (6.70) | NCBInr | gi|356582273 | C4A | Complement C4-A isoform 2 preproprotein * | 337 | 7 | −1.8 | 0.00632 |
| 201 | 189,599 (7.39) | NCBInr | gi|1314244 | C4B | Complement C4B precursor | 812 | 11 | −1.8 | 0.00179 |
| 347 | 52,106 (5.37) | SwissProt | FIBG_HUMAN | FGG | Fibrinogen γ chain * | 174 | 29 | −1.6 | 0.033 |
| 359 | 50,117 (6.40) | NCBInr | gi|12054080 | IGHM | Immunoglobulin heavy chain constant region mu ** | 148 | 11 | −1.5 | 0.042 |
| Less abundant in non-relapsed HL (n = 10) | |||||||||
| 315 | 23,725 (4.93) | NCBInr | gi|112877 | ORM1 | α-1-Acid glycoprotein 1 ** | 569 | 33 | 1.6 | 0.049 |
| 373 | 15,991 (5.52) | SwissProt | TTHY_HUMAN | TTR | Transthyretin * | 275 | 56 | 1.7 | 0.049 |
| 484 | 28,061 (5.27) | NCBInr | NP_001304947.1 ° | APOA1 | Apolipoprotein A-I isoform 1 preproprotein | 7462 | 92 | 1.7 | 0.0062 |
| 326 | 30,759 (5.56) | SwissProt | APOA1_HUMAN | APOA1 | Apolipoprotein A-I ** | 524 | 56 | 2.1 | 0.030 |
| 321 | 53,031(5.89) | SwissProt | CLUS_HUMAN | CLU | Clusterin * | 209 | 17 | 2.2 | 0.019 |
| 340 | 45,371 (5.28) | SwissProt | APOA4_HUMAN | APOA4 | Apolipoprotein A-IV ** | 294 | 44 | 2.3 | 0.018 |
| 335 | 45,371 (5.28) | NCBInr | gi|178759 | APOA4 | Apolipoprotein A-IV | 700 | 61 | 2.3 | 0.014 |
| 333 | 45,371 (5.28) | SwissProt | APOA4_HUMAN | APOA4 | Apolipoprotein A-IV | 314 | 56 | 2.5 | 0.016 |
| 548 | 45,861 (6.13) | SwissProt | HPT_HUMAN | HP | Haptoglobin * | 397 | 10 | 3.2 | 0.00895 |
| 561 | 45,861 (6.13) | SwissProt | HPT_HUMAN | HP | Haptoglobin * | 261 | 10 | 3.3 | 0.017 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Repetto, O.; Lovisa, F.; Elia, C.; Enderle, D.; Romanato, F.; Buffardi, S.; Sala, A.; Pillon, M.; Steffan, A.; Burnelli, R.; et al. Proteomic Exploration of Plasma Exosomes and Other Small Extracellular Vesicles in Pediatric Hodgkin Lymphoma: A Potential Source of Biomarkers for Relapse Occurrence. Diagnostics 2021, 11, 917. https://doi.org/10.3390/diagnostics11060917
Repetto O, Lovisa F, Elia C, Enderle D, Romanato F, Buffardi S, Sala A, Pillon M, Steffan A, Burnelli R, et al. Proteomic Exploration of Plasma Exosomes and Other Small Extracellular Vesicles in Pediatric Hodgkin Lymphoma: A Potential Source of Biomarkers for Relapse Occurrence. Diagnostics. 2021; 11(6):917. https://doi.org/10.3390/diagnostics11060917
Chicago/Turabian StyleRepetto, Ombretta, Federica Lovisa, Caterina Elia, Daniel Enderle, Filippo Romanato, Salvatore Buffardi, Alessandra Sala, Marta Pillon, Agostino Steffan, Roberta Burnelli, and et al. 2021. "Proteomic Exploration of Plasma Exosomes and Other Small Extracellular Vesicles in Pediatric Hodgkin Lymphoma: A Potential Source of Biomarkers for Relapse Occurrence" Diagnostics 11, no. 6: 917. https://doi.org/10.3390/diagnostics11060917








