Effect of Multishell Diffusion MRI Acquisition Strategy and Parcellation Scale on Rich-Club Organization of Human Brain Structural Networks
Abstract
:1. Introduction
2. Material and Methods
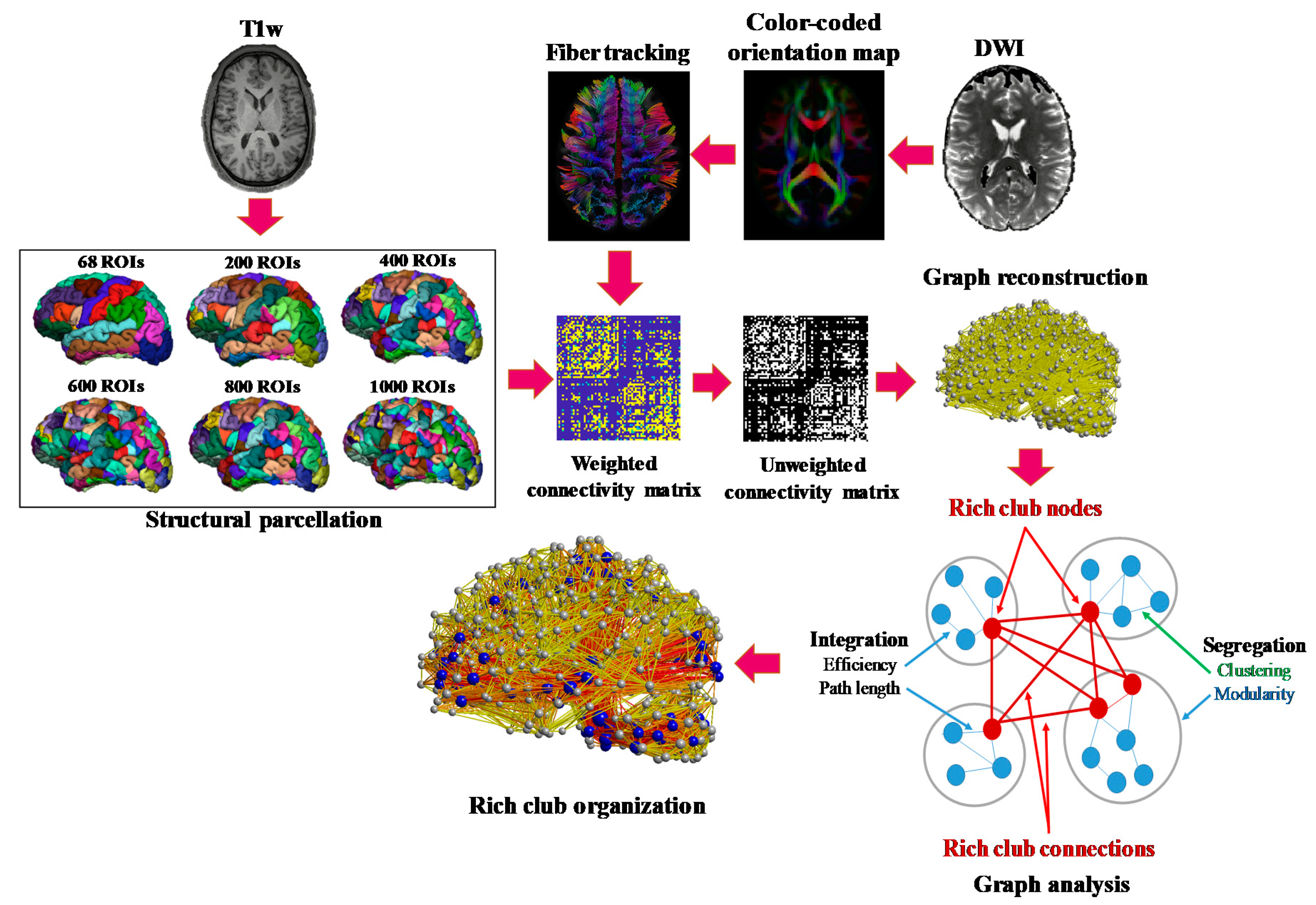
2.1. Processing Pipeline
2.2. Imaging Data and Preprocessing
2.3. Structural Connectome Reconstruction
2.3.1. Network Construction
2.3.2. Topological Properties
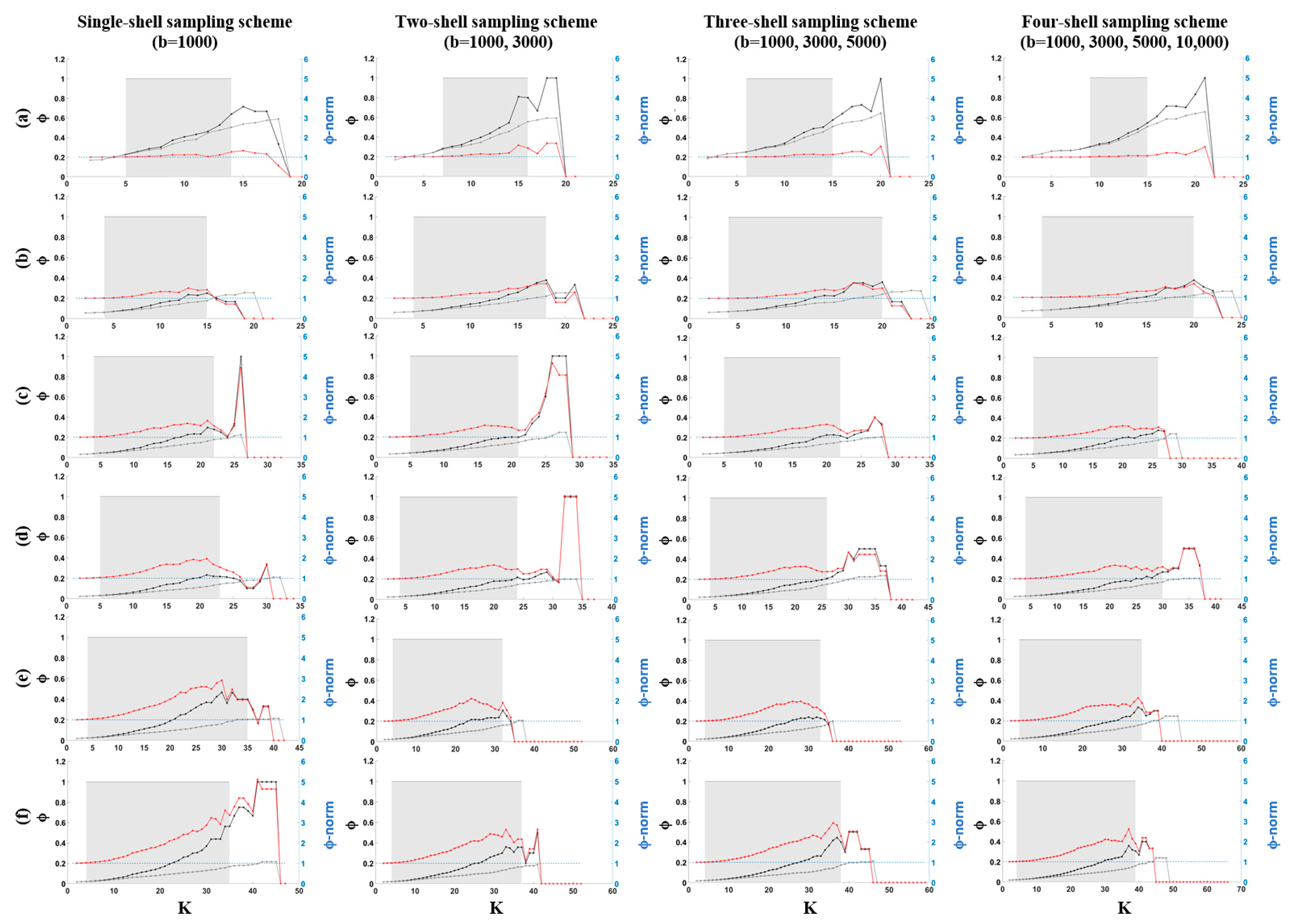
2.3.3. Rich-Club Coefficient for Unweighted Networks
2.4. Statistical Test
3. Results
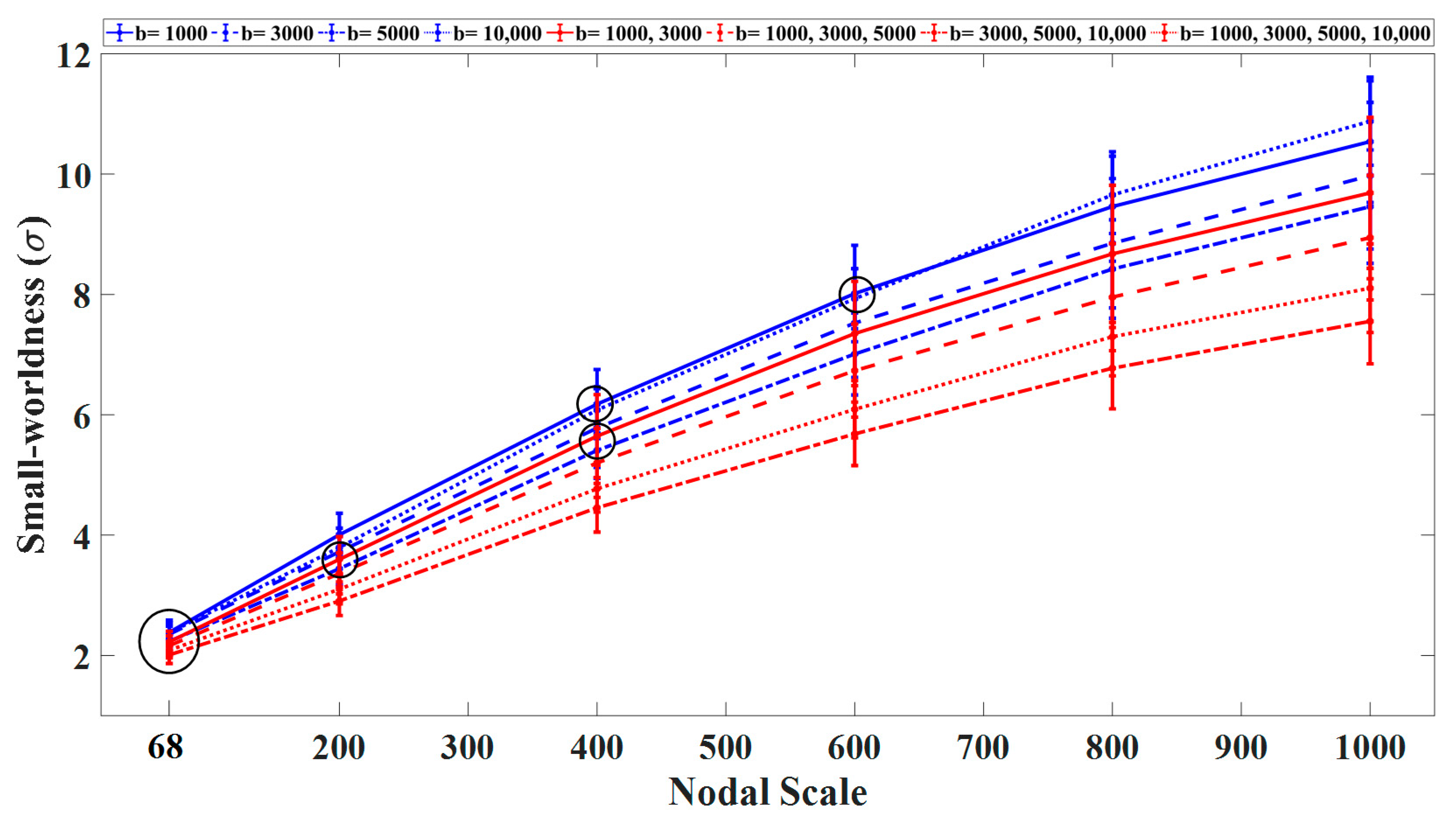
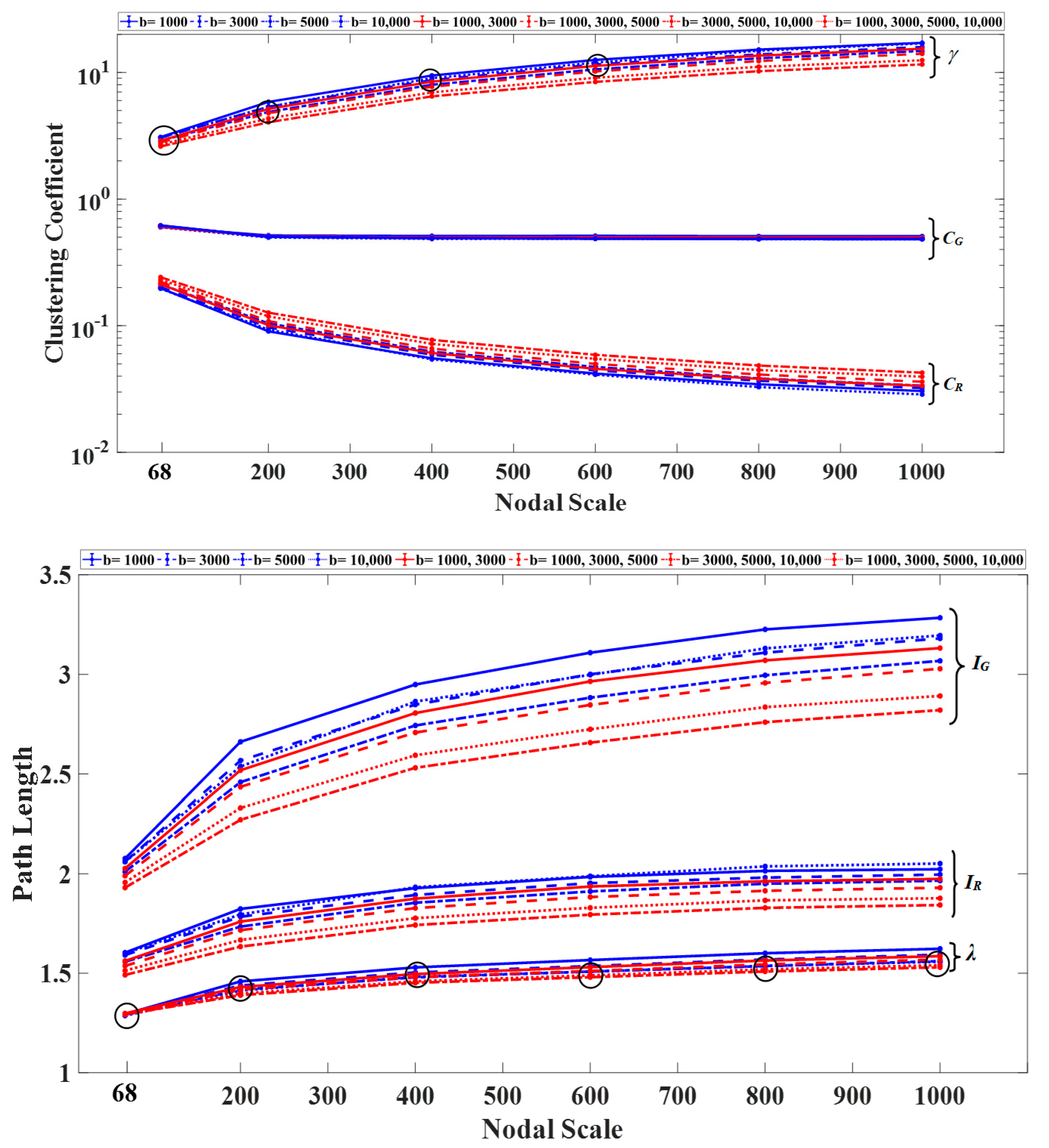
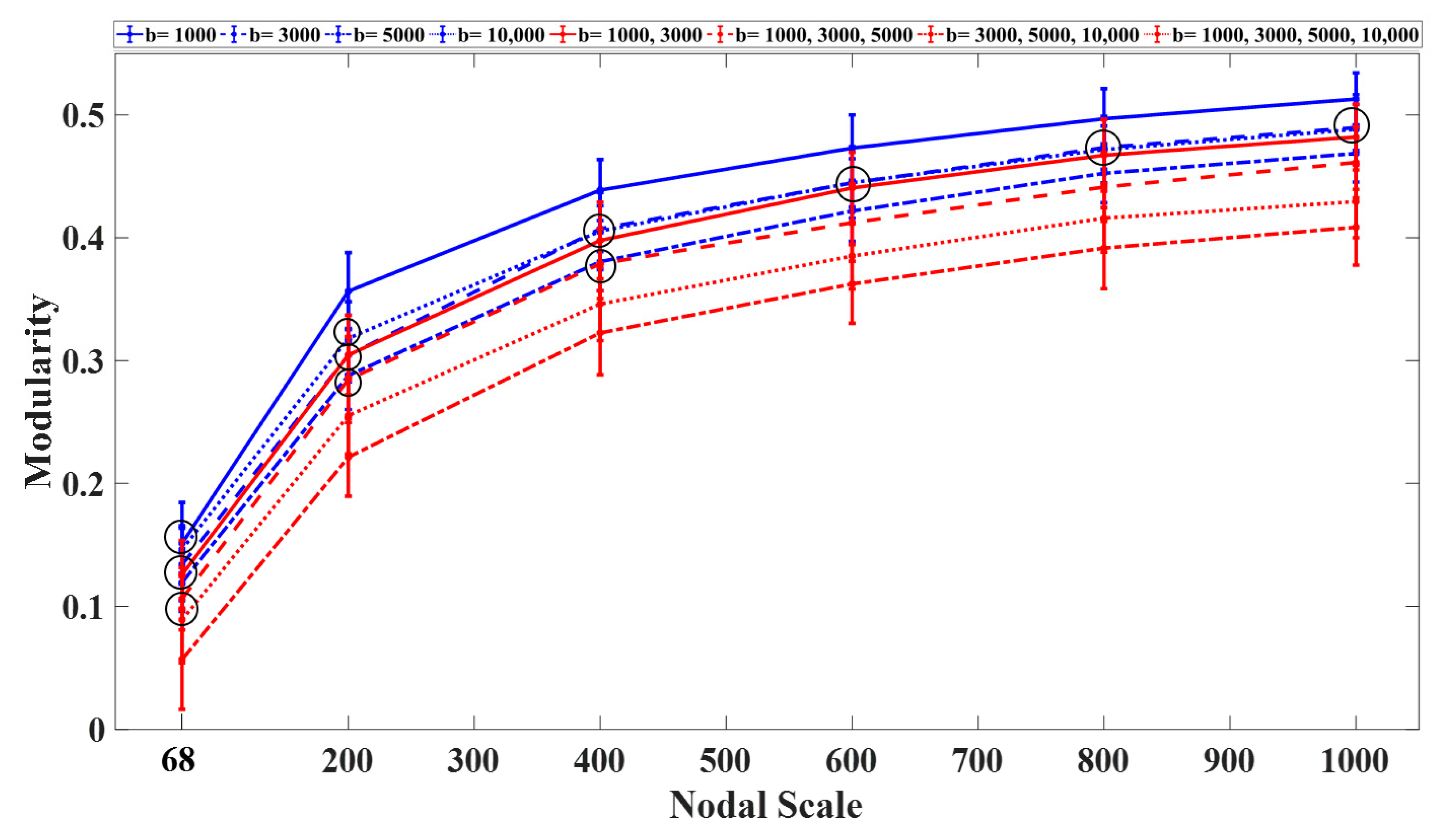
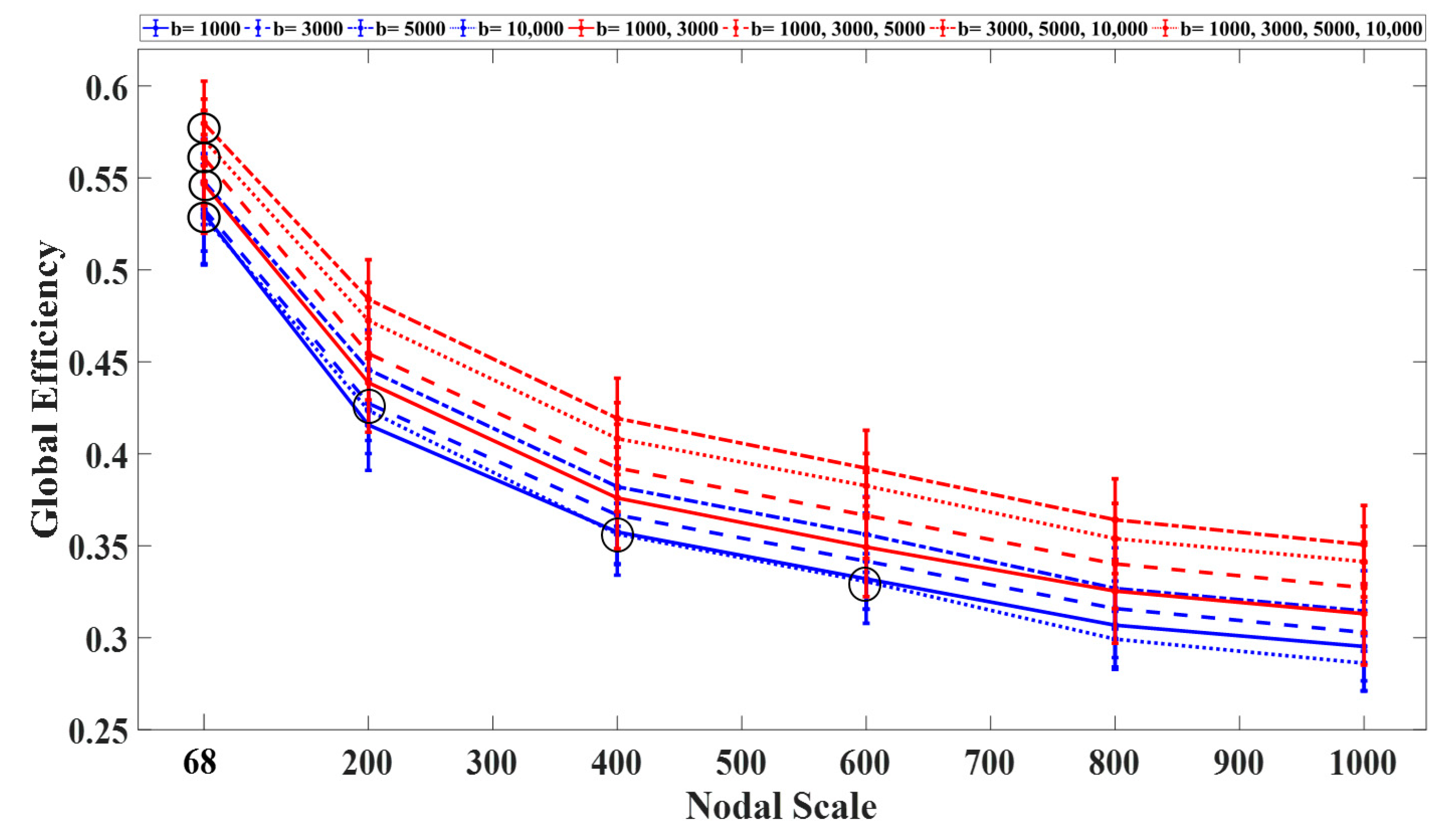
3.1. Graph Measures
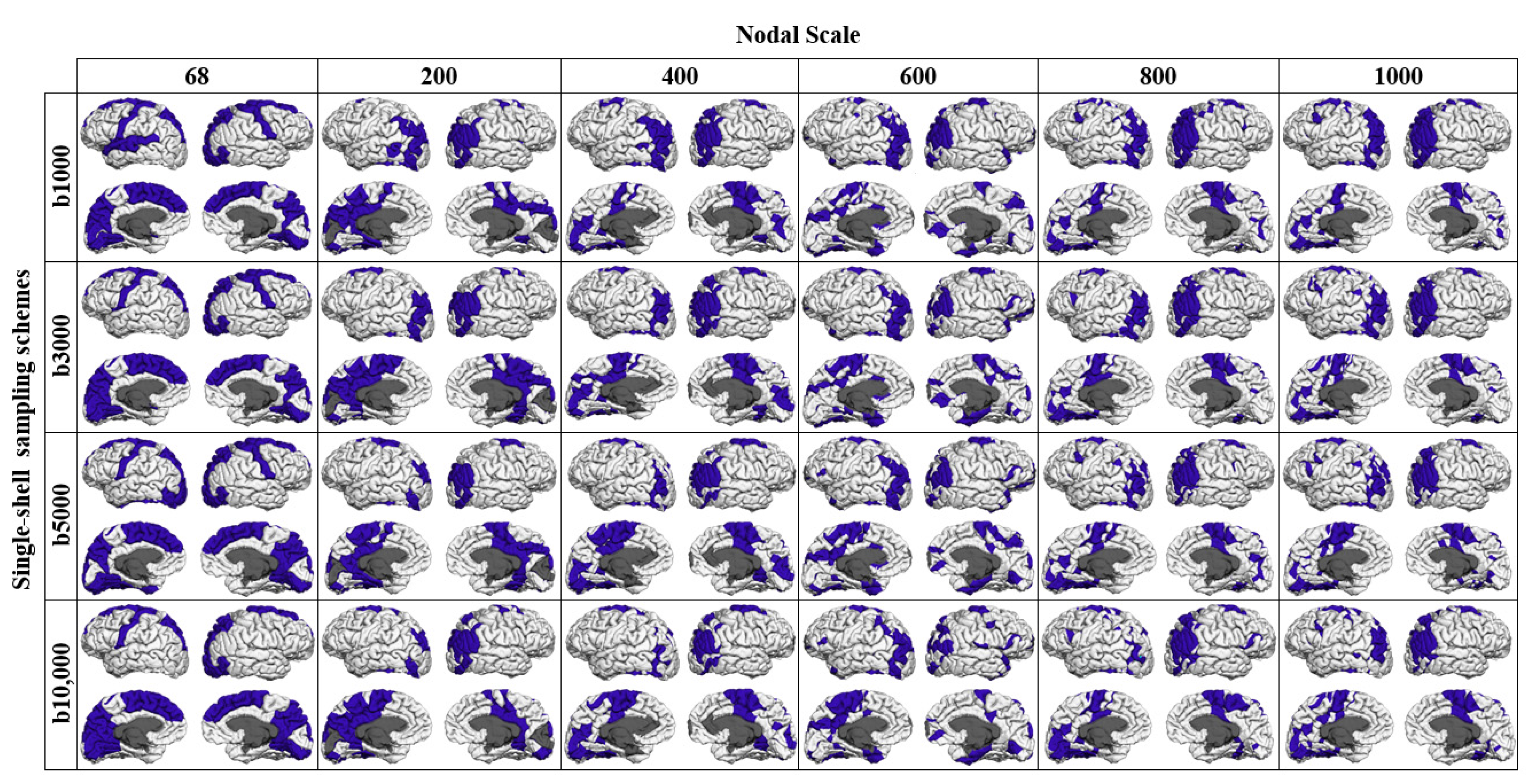
3.2. Rich-Club Organization of Structural Brain Networks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heuvel, M.P.V.D.; Sporns, O. An Anatomical Substrate for Integration among Functional Networks in Human Cortex. J. Neurosci. 2013, 33, 14489–14500. [Google Scholar] [CrossRef] [PubMed]
- Zalesky, A.; Fornito, A.; Harding, I.H.; Cocchi, L.; Yücel, M.; Pantelis, C.; Bullmore, E.T. Whole-brain anatomical networks: Does the choice of nodes matter? NeuroImage 2010, 50, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Cammoun, L.; Gigandet, X.; Meskaldji, D.; Thiran, J.P.; Sporns, O.; Do, K.Q.; Maeder, P.; Meuli, R.; Hagmann, P. Mapping the human connectome at multiple scales with diffusion spectrum MRI. J. Neurosci. Methods 2012, 203, 386–397. [Google Scholar] [CrossRef] [Green Version]
- Sporns, O.; Tononi, G.; Kötter, R. The Human Connectome: A Structural Description of the Human Brain. PLoS Comput. Biol. 2005, 1, e42. [Google Scholar] [CrossRef]
- De Reus, M.A.; Heuvel, M.P.V.D. Rich Club Organization and Intermodule Communication in the Cat Connectome. J. Neurosci. 2013, 33, 12929–12939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.; Fias, W.; Caeyenberghs, K.; Marinazzo, D. Brain networks under attack: Robustness properties and the impact of lesions. Brain 2016, 139 Pt 12, 3063–3083. [Google Scholar] [CrossRef]
- Gratton, C.; Nomura, E.M.; Pérez, F.; D’Esposito, M. Focal Brain Lesions to Critical Locations Cause Widespread Disruption of the Modular Organization of the Brain. J. Cogn. Neurosci. 2012, 24, 1275–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-J.; Min, B.-K. Rich-club in the brain’s macrostructure: Insights from graph theoretical analysis. Comput. Struct. Biotechnol. J. 2020, 18, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Wang, W.; Yang, L.; Chen, K.; Chen, R.; Han, Y. Rich club disturbances of the human connectome from subjective cognitive decline to Alzheimer’s disease. Theranostics 2018, 8, 3237–3255. [Google Scholar] [CrossRef]
- Bullmore, E.T.; Sporns, O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef]
- Betzel, R.F.; Bassett, D.S. Multi-scale brain networks. NeuroImage 2017, 160, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosey, T.; Williams, G.; Ansorge, R. Inference of multiple fiber orientations in high angular resolution diffusion imaging. Magn. Reson. Med. 2005, 54, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Frank, L.R. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magn. Reson. Med. 2002, 47, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Witzel, T.; Nummenmaa, A.; Van Dijk, K.R.; Van Horn, J.D.; Drews, M.K.; Somerville, L.H.; Sheridan, M.A.; Santillana, R.M.; Snyder, J.; et al. MGH–USC Human Connectome Project datasets with ultra-high b-value diffusion MRI. NeuroImage 2016, 124 Pt B, 1108–1114. [Google Scholar] [CrossRef] [Green Version]
- Heuvel, M.P.V.D.; Kahn, R.S.; Goñi, J.; Sporns, O. High-cost, high-capacity backbone for global brain communication. Proc. Natl. Acad. Sci. USA 2012, 109, 11372–11377. [Google Scholar] [CrossRef] [Green Version]
- Andersson, J.L.; Skare, S. A Model-Based Method for Retrospective Correction of Geometric Distortions in Diffusion-Weighted EPI. NeuroImage 2002, 16, 177–199. [Google Scholar] [CrossRef]
- Greve, D.N.; Fischl, B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage 2009, 48, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Yeh, F.-C.; Wedeen, V.J.; Tseng, W.-Y.I. Generalized q-Sampling Imaging. IEEE Trans. Med. Imaging 2010, 29, 1626–1635. [Google Scholar] [CrossRef]
- Yeh, F.-C.; Verstynen, T.D.; Wang, Y.; Fernández-Miranda, J.C.; Tseng, W.-Y.I. Deterministic Diffusion Fiber Tracking Improved by Quantitative Anisotropy. PLoS ONE 2013, 8, e80713. [Google Scholar] [CrossRef] [Green Version]
- Maffei, C.; Sarubbo, S.; Jovicich, J. Diffusion-based tractography atlas of the human acoustic radiation. Sci. Rep. 2019, 9, 4046. [Google Scholar] [CrossRef] [Green Version]
- Yeh, F.-C.; Zaydan, I.M.; Suski, V.R.; Lacomis, D.; Richardson, R.M.; Maroon, J.C.; Barrios-Martinez, J. Differential tractography as a track-based biomarker for neuronal injury. NeuroImage 2019, 202, 116131. [Google Scholar] [CrossRef]
- Yeh, F.-C.; Panesar, S.; Barrios, J.; Fernandes, D.; Abhinav, K.; Meola, A.; Fernandez-Miranda, J.C. Automatic Removal of False Connections in Diffusion MRI Tractography Using Topology-Informed Pruning (TIP). Neurotherapeutics 2019, 16, 52–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 2006, 31, 968–980. [Google Scholar] [CrossRef]
- Jenkinson, M.; Bannister, P.; Brady, M.; Smith, S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 2002, 17, 825–841. [Google Scholar] [CrossRef] [PubMed]
- Heuvel, M.P.V.D.; Sporns, O. Rich-Club Organization of the Human Connectome. J. Neurosci. 2011, 31, 15775–15786. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassett, D.S.; Bullmore, E.T. Human brain networks in health and disease. Curr. Opin. Neurol. 2009, 22, 340–347. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Bao, Y.; Wang, Y.; Li, Z.; Zheng, X.; Long, S.; Wang, Y. Differences between generalized Q-sampling imaging and diffusion tensor imaging in visualization of crossing neural fibers in the brain. Surg. Radiol. Anat. 2019, 41, 1019–1028. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Cieslak, M.; Greene, C.; Grafton, S.T.; Carlson, J.M. Sensitivity analysis of human brain structural network construction. Netw. Neurosci. 2017, 1, 446–467. [Google Scholar] [CrossRef]
- Xie, S.; Zuo, N.; Shang, L.; Song, M.; Fan, L.; Jiang, T. How Does B-Value Affect HARDI Reconstruction Using Clinical Diffusion MRI Data? PLoS ONE 2015, 10, e0120773. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, P.; Prats-Galino, A.; Gallardo-Pujol, D.; Villoslada, P.; Falcón, C.; Prčkovska, V. Evaluating Structural Connectomics in Relation to Different Q-space Sampling Techniques. MICCAI Int. Conf. Med. Image Comput. Comput. Assist. Interv. 2013, 16 Pt 1, 671–678. [Google Scholar] [CrossRef]
- Bullmore, E.T.; Sporns, O. The economy of brain network organization. Nat. Rev. Neurosci. 2012, 13, 336–349. [Google Scholar] [CrossRef] [PubMed]
- Griffa, A.; Heuvel, M.P.V.D. Rich-club neurocircuitry: Function, evolution, and vulnerability. Dialog Clin. Neurosci. 2018, 20, 121–132. [Google Scholar]
- Sarwar, T.; Ramamohanarao, K.; Zalesky, A. Mapping connectomes with diffusion MRI: Deterministic or probabilistic tractography? Magn. Reson. Med. 2019, 81, 1368–1384. [Google Scholar] [CrossRef]
- Zalesky, A.; Fornito, A.; Cocchi, L.; Gollo, L.L.; Heuvel, M.P.V.D.; Breakspear, M. Connectome sensitivity or specificity: Which is more important? NeuroImage 2016, 142, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Calamante, F. The Seven Deadly Sins of Measuring Brain Structural Connectivity Using Diffusion MRI Streamlines Fibre-Tracking. Diagnostics 2019, 9, 115. [Google Scholar] [CrossRef] [Green Version]
- Morgan, P.S.; Bowtell, R.W.; McIntyre, D.J.; Worthington, B.S. Correction of spatial distortion in EPI due to inhomo-geneous static magnetic fields using the reversed gradient method. J. Magn. Reson. Imaging 2004, 19, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Cheryauka, A.B.; Lee, J.N.; Samsonov, A.A.; Defrise, M.; Gullberg, G.T. MRI diffusion tensor reconstruction with PROPELLER data acquisition. Magn. Reson. Imaging 2004, 22, 139–148. [Google Scholar] [CrossRef]
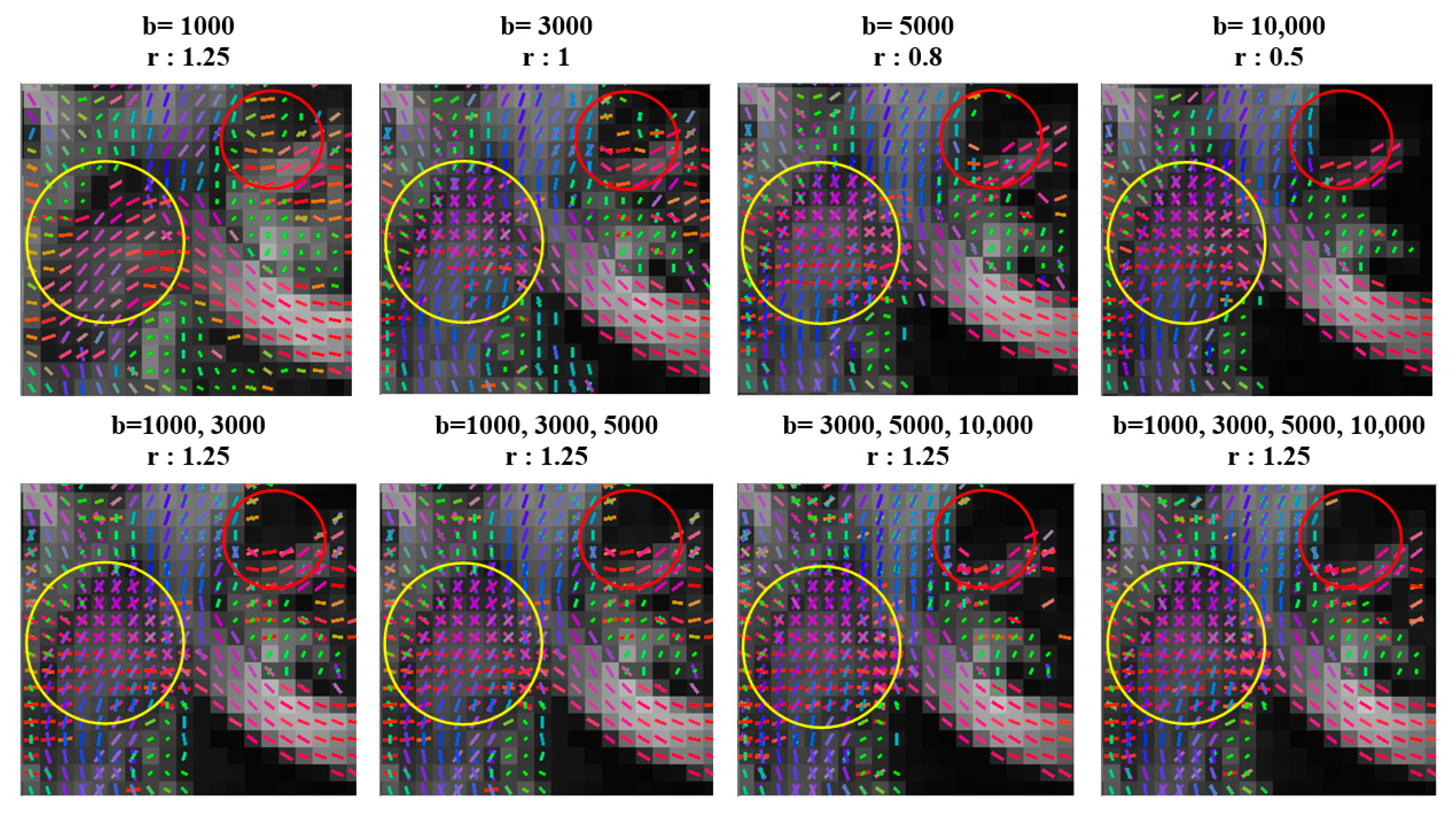

| Single-Shell Sampling Schemes | Multishell Sampling Schemes | |||||||
|---|---|---|---|---|---|---|---|---|
| b = 1000 | b = 3000 | b = 5000 | b = 10,000 | b = 1000, 3000 | b = 1000, 3000, 5000 | b = 3000, 5000, 10,000 | b = 1000, 3000, 5000, 10,000 | |
| SL (mm) (mean ± S.D.) | 65.6 ± 2.9 | 66.9 ± 2.6 | 69.2 ± 2.8 | 69.9 ± 1.9 | 66.9 ± 2.9 | 68.6 ± 2.7 | 70.1 ± 2.8 | 69.5 ± 2.6 |
| nQA (a.u.) (mean ± S.D.) | 0.142 ± 0.028 | 0.123 ± 0.017 | 0.112 ± 0.011 | 0.104 ± 0.022 | 0.125 ± 0.019 | 0.116 ± 0.013 | 0.106 ± 0.009 | 0.109 ± 0.009 |
| Nodal Scale | ||||||||
|---|---|---|---|---|---|---|---|---|
| 68 | 200 | 400 | 600 | 800 | 1000 | |||
| Single-shell sampling schemes | b = 1000 | k level | 12 | 12 | 14 | 14 | 15 | 15 |
| RC | 15.2 | 17.2 | 25.6 | 29.9 | 35.9 | 40.4 | ||
| FC | 48.1 | 39.2 | 33.6 | 29.2 | 26.6 | 22.8 | ||
| LC | 36.7 | 43.6 | 40.8 | 40.9 | 37.4 | 36.8 | ||
| b = 3000 | k level | 13 | 14 | 15 | 16 | 16 | 17 | |
| RC | 15.9 | 20.2 | 25.2 | 28.2 | 34.3 | 36.8 | ||
| FC | 45.3 | 33.6 | 32.2 | 31.9 | 27.1 | 26.5 | ||
| LC | 38.9 | 46.2 | 42.5 | 39.8 | 38.6 | 36.6 | ||
| b = 5000 | k level | 15 | 15 | 16 | 16 | 16 | 17 | |
| RC | 13.3 | 19.6 | 24.5 | 26.7 | 31.4 | 35.1 | ||
| FC | 50 | 33.9 | 32.1 | 33.2 | 30.8 | 26.9 | ||
| LC | 36.6 | 46.5 | 43.4 | 40.1 | 37.8 | 37.9 | ||
| b = 10,000 | k level | 14 | 14 | 14 | 13 | 14 | 14 | |
| RC | 18.5 | 19.2 | 25.4 | 27.9 | 34.7 | 37.9 | ||
| FC | 40.6 | 35.7 | 34.3 | 33.5 | 29.9 | 27.9 | ||
| LC | 40.9 | 45.1 | 40.3 | 38.5 | 35.4 | 34.1 | ||
| Multishell sampling schemes | b = 1000, | k level | 14 | 14 | 15 | 16 | 17 | 17 |
| 3000 | RC | 14.5 | 19.6 | 23.7 | 28 | 33.3 | 36.9 | |
| FC | 45.2 | 33.9 | 33 | 31.6 | 28.3 | 26.2 | ||
| LC | 40.3 | 46.5 | 43.3 | 40.4 | 38.4 | 36.9 | ||
| b = 1000, 3000, | k level | 15 | 16 | 17 | 17 | 18 | 19 | |
| 5000 | RC | 14.7 | 17.8 | 22.6 | 25.1 | 30.7 | 34.2 | |
| FC | 45.2 | 36.8 | 34.1 | 34.1 | 30 | 27.5 | ||
| LC | 40.1 | 45.3 | 43.3 | 40.8 | 39.3 | 38.3 | ||
| b = 3000, 5000, | k level | 15 | 18 | 18 | 19 | 21 | 22 | |
| 10,000 | RC | 13.3 | 16.8 | 21.4 | 25.4 | 30.5 | 32.8 | |
| FC | 45.8 | 38.3 | 35.1 | 32.6 | 28.9 | 28 | ||
| LC | 40.9 | 44.9 | 43.5 | 42 | 40.6 | 39.2 | ||
| b = 1000, 3000, | k level | 15 | 17 | 18 | 18 | 19 | 20 | |
| 5000, 10,000 | RC | 12.9 | 17.4 | 22.5 | 24.3 | 30.4 | 33.3 | |
| FC | 46.9 | 35.5 | 32.7 | 34.7 | 29.3 | 27.9 | ||
| LC | 40.2 | 47.1 | 44.7 | 41.1 | 40.4 | 38.8 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalilian, M.; Kazemi, K.; Fouladivanda, M.; Makki, M.; Helfroush, M.S.; Aarabi, A. Effect of Multishell Diffusion MRI Acquisition Strategy and Parcellation Scale on Rich-Club Organization of Human Brain Structural Networks. Diagnostics 2021, 11, 970. https://doi.org/10.3390/diagnostics11060970
Khalilian M, Kazemi K, Fouladivanda M, Makki M, Helfroush MS, Aarabi A. Effect of Multishell Diffusion MRI Acquisition Strategy and Parcellation Scale on Rich-Club Organization of Human Brain Structural Networks. Diagnostics. 2021; 11(6):970. https://doi.org/10.3390/diagnostics11060970
Chicago/Turabian StyleKhalilian, Maedeh, Kamran Kazemi, Mahshid Fouladivanda, Malek Makki, Mohammad Sadegh Helfroush, and Ardalan Aarabi. 2021. "Effect of Multishell Diffusion MRI Acquisition Strategy and Parcellation Scale on Rich-Club Organization of Human Brain Structural Networks" Diagnostics 11, no. 6: 970. https://doi.org/10.3390/diagnostics11060970







