Strongyloides-Specific IgE Phage cDNA Clones and Development of a Novel ELISA for Strongyloidiasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Serum Samples
2.2. Serum Pre-Adsorption for Immunoscreening
2.3. IgG-Enzyme Linked Immunosorbent Assay (ELISA) to Check the Efficiency of Serum Pre-Adsorption
2.4. Immunoscreening of S. stercoralis Complementary DNA (cDNA) Library
2.5. Post-Immunoscreening Analysis
2.6. Custom Cloning into a pET32a Expression System
2.7. Expression and Purification of Recombinant Protein
2.8. Western Blot Analysis
2.9. Confirmation of Protein Identity by LC-MS-MS
2.10. Development of an IgE-ELISA Using the Recombinant Protein
2.11. Statistical Analysis
3. Results
3.1. Serum Pre-Adsorption
3.2. Immunoscreening of S. stercoralis cDNA Library
3.3. Sequence Analysis
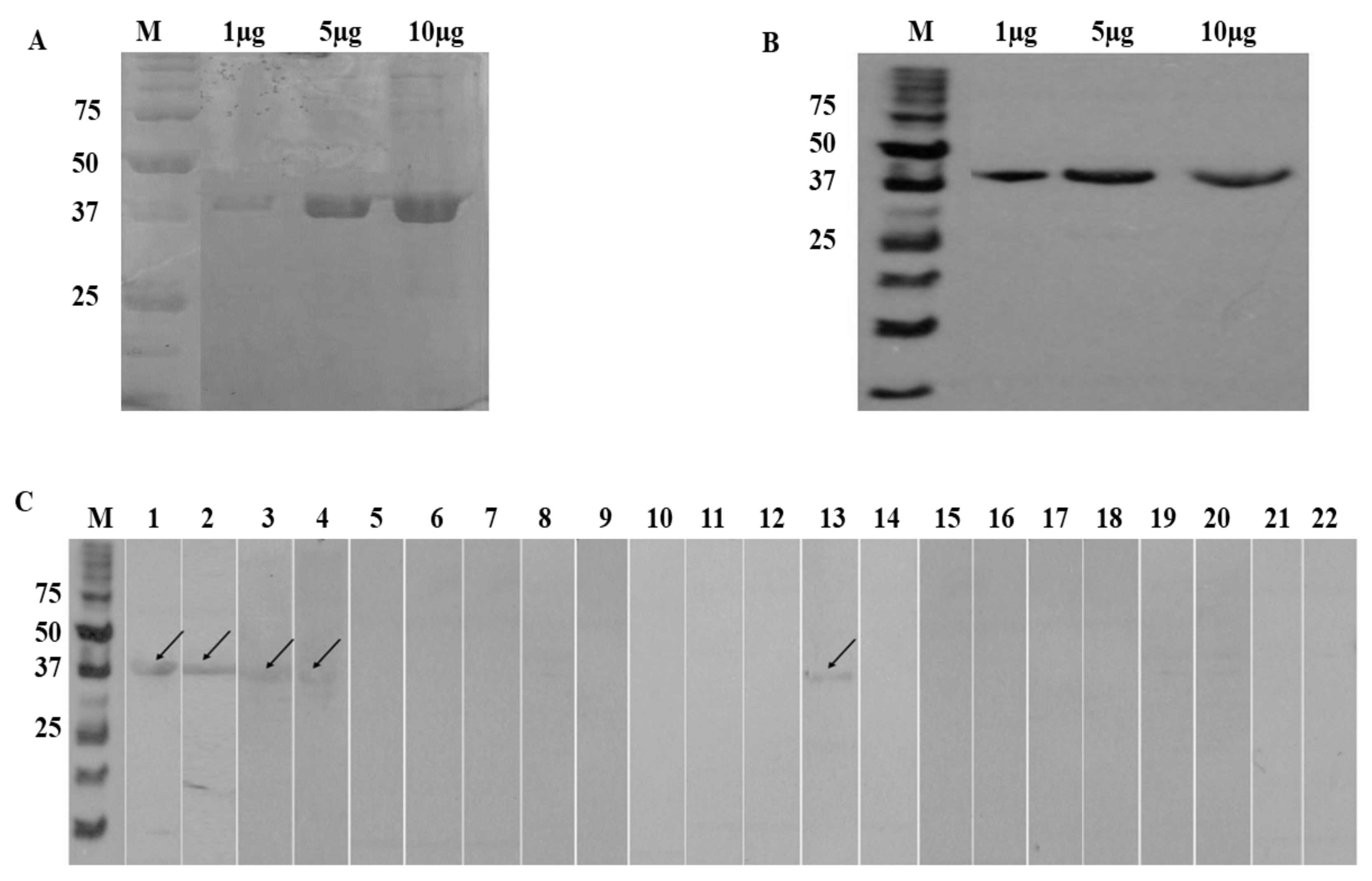
3.4. SDS-PAGE and Western Blot
3.5. LC-MS-MS
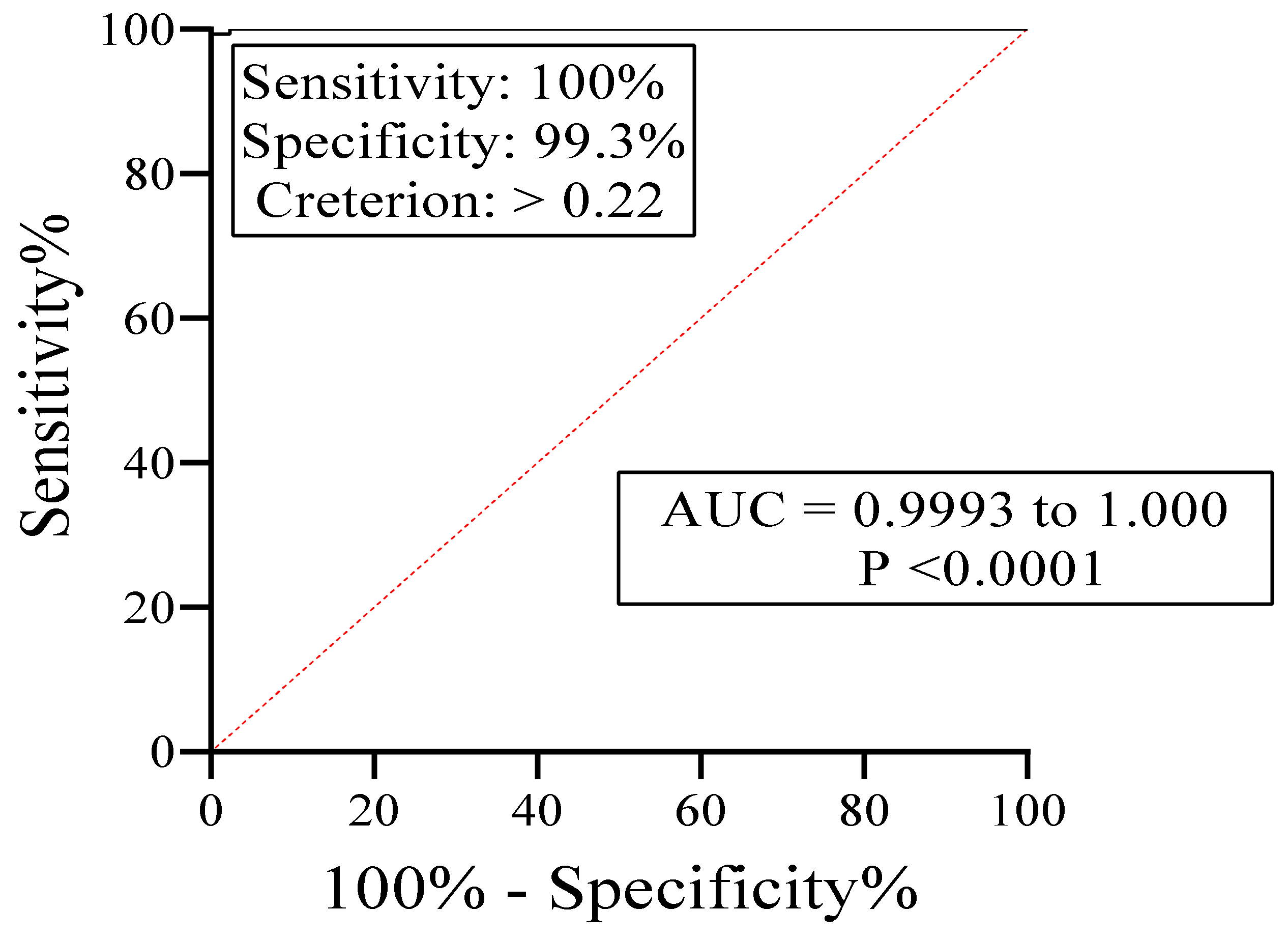
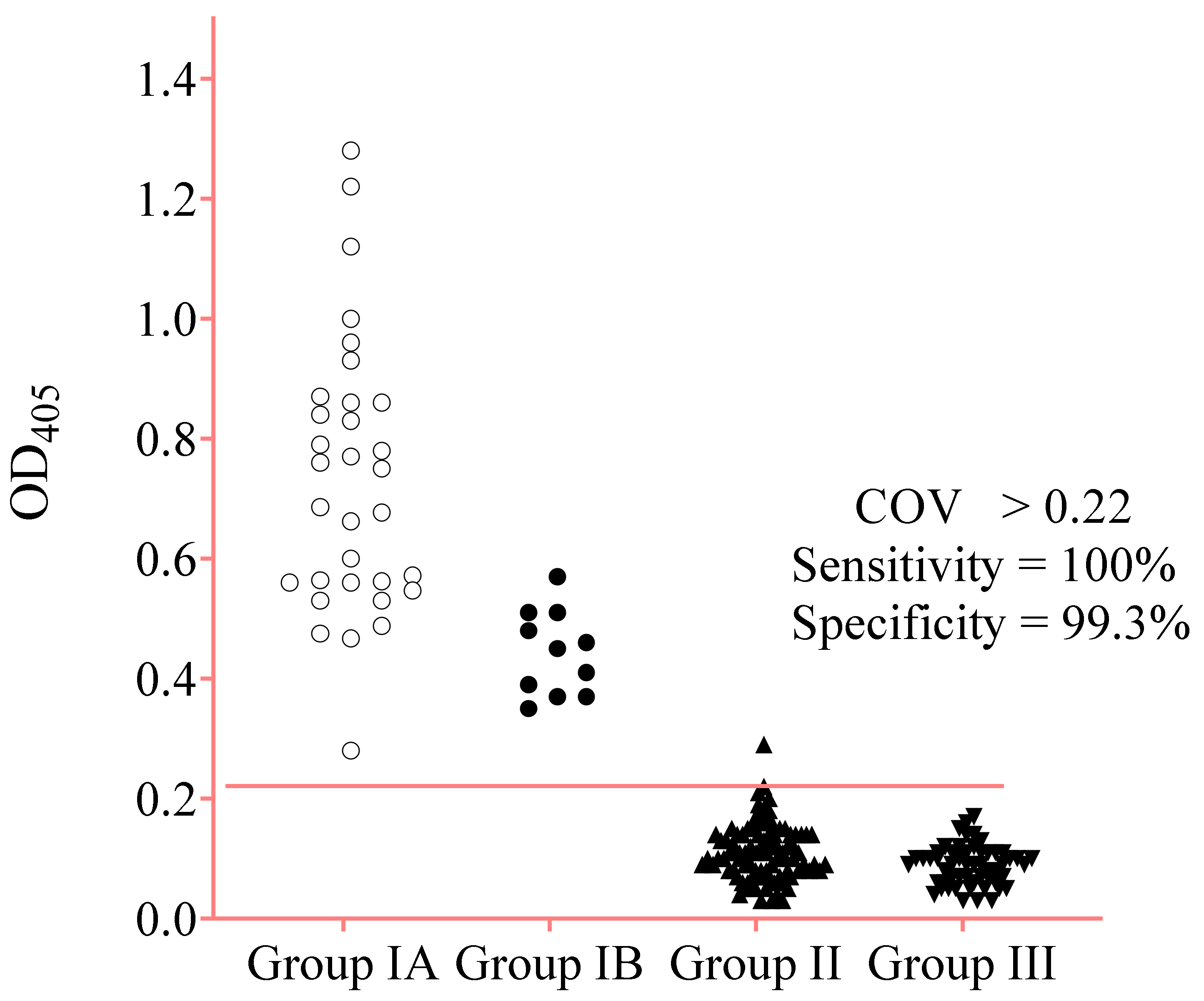
3.6. IgE-ELISA Using rA133
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schad, G.A. Morphology and life history of Strongyloides stercoralis. In Strongyloidiasis a Major Roundworm Infection of Man; Grove, D.I., Ed.; Taylor & Francis: London, UK, 1989; pp. 85–104. [Google Scholar]
- Thanchomnang, T.; Intapan, P.M.; Sanpool, O.; Rodpai, R.; Tourtip, S.; Yahom, S.; Kullawat, J.; Radomyos, P.; Thammasiri, C.; Maleewong, W. First molecular identification and genetic diversity of Strongyloides stercoralis and Strongyloides fuelleborni in human communities having contact with long-tailed macaques in Thailand. Parasitol. Res. 2017, 116, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Sheild, J.M.; Kow, F.; Shield, J.M.; Kow, F. A comparative study of intestinal helminths in pre-school age urban and rural children in Morobe Province, Papua New Guinea. Papua New Guinea Med. J. 2013, 56, 14–31. [Google Scholar]
- Bisoffi, Z.; Buonfrate, D.; Montresor, A.; Requena-Méndez, A.; Munoz, J.; Krolewiecki, A.J.; Gotuzzo, E.; Mena, M.A.; Chiodini, P.L.; Anselmi, M. Strongyloides stercoralis: A plea for action. PLoS Negl. Trop. Dis. 2013, 7, e2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schar, F.; Trostdorf, U.; Giardina, F.; Khieu, V.; Muth, S.; Marti, H.; Vounatsou, P.; Odermatt, P. Strongyloides stercoralis: Global Distribution and Risk Factors. PLoS Negl. Trop. Dis. 2013, 7, e2288. [Google Scholar] [CrossRef] [Green Version]
- Utzinger, J.; Hatz, C.; Adjossan, L.; Sieto, B.; Silué, K.D.; Becker, S.L.; Kern, W.V.; Koné, S.; N’Goran, E.K. Diagnosis, Clinical Features, and Self-Reported Morbidity of Strongyloides stercoralis and Hookworm Infection in a Co-Endemic Setting. PLoS Negl. Trop. Dis. 2011, 5, e1292. [Google Scholar] [CrossRef] [Green Version]
- Mejia, R.; Nutman, T.B. Screening, prevention, and treatment for hyperinfection syndrome and disseminated infections caused by Strongyloides stercoralis. Curr. Opin. Infect. Dis. 2012, 25, 458–463. [Google Scholar] [CrossRef] [Green Version]
- Keiser, P.B.; Nutman, T.B. Strongyloides stercoralis in the Immunocompromised Population. Clin. Microbiol. Rev. 2004, 17, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Puthiyakunnon, S.; Boddu, S.; Li, Y.; Zhou, X.; Wang, C.; Li, J.; Chen, X. Strongyloidiasis—An insight into its global prevalence and management. PLoS Negl. Trop. Dis. 2014, 8, e3018. [Google Scholar] [CrossRef] [Green Version]
- Lam, C.S.; Tong, M.K.H.; Chan, K.M.; Siu, Y.P. Disseminated strongyloidiasis: A retrospective study of clinical course and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 14–18. [Google Scholar] [CrossRef]
- Buonfrate, D.; Sequi, M.; Mejia, R.; Cimino, R.O.; Krolewiecki, A.J.; Albonico, M.; Degani, M.; Tais, S.; Angheben, A.; Requena-Mendez, A. Accuracy of five serologic tests for the follow up of Strongyloides stercoralis infection. PLoS Negl. Trop. Dis. 2015, 9, e0003491. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, E.F.; Terashima, A.; Pineda-Reyes, J.; Vasquez-Rios, G.; Marin, R.; Pineda-Reyes, R. Strongyloides stercoralis hyperinfection syndrome: A deeper understanding of a neglected disease. J. Parasit. Dis. 2019, 43, 167–175. [Google Scholar] [CrossRef]
- Schaffel, R.; Nucci, M.; Carvalho, E.; Braga, M.; Almeida, L.; Portugal, R.; Pulcheri, W. The value of an immunoenzymatic test (enzyme-linked immunosorbent assay) for the diagnosis of strongyloidiasis in patients immunosuppressed by hematologic malignancies. Am. J. Trop. Med. Hyg. 2001, 65, 346–350. [Google Scholar] [CrossRef] [Green Version]
- Genta, R.M.; Schad, G.A.; Hellman, M.E. Strongyloides stercoralis: Parasitological, immunological and pathological observations in immunosuppressed dogs. Trans. R. Soc. Trop. Med. Hyg. 1986, 80, 34–41. [Google Scholar] [CrossRef]
- Ming, D.K.; Armstrong, M.; Lowe, P.; Chiodini, P.L.; Doherty, J.F.; Whitty, C.J.M.; McGregor, A.C. Clinical and Diagnostic Features of 413 Patients Treated for Imported Strongyloidiasis at the Hospital for Tropical Diseases, London. Am. J. Trop. Med. Hyg. 2019, 101, 428–431. [Google Scholar] [CrossRef]
- Bisoffi, Z.; Buonfrate, D.; Sequi, M.; Mejia, R.; Cimino, R.O.; Krolewiecki, A.J.; Albonico, M.; Gobbo, M.; Bonafini, S.; Angheben, A.; et al. Diagnostic Accuracy of Five Serologic Tests for Strongyloides stercoralis Infection. PLoS Negl. Trop. Dis. 2014, 8, e2640. [Google Scholar] [CrossRef] [Green Version]
- Loutfy, M.R.; Wilson, M.; Keystone, J.S.; Kain, K.C. Serology and eosinophil count in the diagnosis and management of strongyloidiasis in a non-endemic area. Am. J. Trop. Med. Hyg. 2002, 66, 749–752. [Google Scholar] [CrossRef] [Green Version]
- Conway, D.J.; Atkins, N.S.; Lillywhite, J.E.; Bailey, J.W.; Robinson, R.D.; Lindo, J.F.; Bundy, D.A.P.; Bianco, A.E. Immunodiagnosis of Strongyloides stercoralis infection: A method for increasing the specificity of the indirect ELISA. Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 173–176. [Google Scholar] [CrossRef]
- Van Dam, G.J.; Stelma, F.F.; Gryseels, B.; Falcão Ferreira, S.T.M.; Talla, I.; Niang, M.; Rotmans, J.P.; Deelder, A.M. Antibody response patterns against Schistosoma mansoni in a recently exposed community in Senegal. J. Infect. Dis. 1996, 173, 1232–1241. [Google Scholar] [CrossRef] [Green Version]
- Ramanathan, R.; Burbelo, P.D.; Groot, S.; Iadarola, M.J.; Neva, F.A.; Nutman, T.B. A Luciferase Immunoprecipitation Systems Assay Enhances the Sensitivity and Specificity of Diagnosis of Strongyloides stercoralis Infection. J. Infect. Dis. 2008, 198, 444–451. [Google Scholar] [CrossRef] [Green Version]
- Norsyahida, A.; Riazi, M.; Sadjjadi, S.M.; Muhammad Hafiznur, Y.; Low, H.C.; Zeehaida, M.; Noordin, R. Laboratory detection of strongyloidiasis: IgG, IgG 4 and IgE-ELISA s and cross-reactivity with lymphatic filariasis. Parasite Immunol. 2013, 35, 174–179. [Google Scholar] [CrossRef]
- Lindo, J.F.; Lee, M.G. Strongyloides stercoralis and S. fulleborni. In Principles and Practice of Clinical Parasitology; Gillespie, S., Pearson, R.D., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2001; pp. 479–500. [Google Scholar]
- Bosqui, L.R.; Corral, M.A.; Levy, D.; Bydlowski, S.P.; Gryschek, R.C.B.; Custodio, L.A.; Pavanelli, W.R.; Conchon-Costa, I.; Costa-Cruz, J.M.; de Paula, F.M. Evaluation of the Dot-ELISA as a diagnostic test for human strongyloidiasis based on the detection of IgA in saliva. Acta Trop. 2020, 203, 105305. [Google Scholar] [CrossRef] [PubMed]
- Bosqui, L.R.; Gonçalves, A.L.R.; Maria do Rosário, F.; Custodio, L.A.; de Menezes, M.C.N.D.; Murad, V.A.; de Paula, F.M.; Pavanelli, W.R.; Conchon-Costa, I.; Costa-Cruz, J.M. Detection of parasite-specific IgG and IgA in paired serum and saliva samples for diagnosis of human strongyloidiasis in northern Paraná state, Brazil. Acta Trop. 2015, 150, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Babu, S.; Nutman, T.B. Immune responses to helminth infection. In Clinical Immunology, 5th ed.; Robert, R., Thomas, F., William, S., Harry, S., Anthony, F., Cornelia, W., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 437–447. [Google Scholar]
- King, C.L.; Xianli, J.; Malhotra, I.; Liu, S.; Mahmoud, A.A.; Oettgen, H.C. Mice with a targeted deletion of the IgE gene have increased worm burdens and reduced granulomatous inflammation following primary infection with Schistosoma mansoni. J. Immunol. 1997, 158, 294–300. [Google Scholar] [PubMed]
- Spencer, L.; Shultz, L.; Rajan, T. V Interleukin-4 receptor–Stat6 signaling in murine infections with a tissue-dwelling nematode parasite. Infect. Immun. 2001, 69, 7743–7752. [Google Scholar] [CrossRef] [Green Version]
- Gurish, M.F.; Bryce, P.J.; Tao, H.; Kisselgof, A.B.; Thornton, E.M.; Miller, H.R.; Friend, D.S.; Oettgen, H.C. IgE enhances parasite clearance and regulates mast cell responses in mice infected with Trichinella spiralis. J. Immunol. 2004, 172, 1139–1145. [Google Scholar] [CrossRef] [Green Version]
- Rihet, P.; Demeure, C.E.; Bourgois, A.; Prata, A.; Dessein, A.J. Evidence for an association between human resistance to Schistosoma mansoni and high anti-larval IgE levels. Eur. J. Immunol. 1991, 21, 2679–2686. [Google Scholar] [CrossRef]
- Faulkner, H.; Turner, J.; Kamgno, J.; Pion, S.D.; Boussinesq, M.; Bradley, J.E. Age-and infection intensity-dependent cytokine and antibody production in human trichuriasis: The importance of IgE. J. Infect. Dis. 2002, 185, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Bethony, J.; Loukas, A.; Smout, M.; Brooker, S.; Mendez, S.; Plieskatt, J.; Goud, G.; Bottazzi, M.E.; Zhan, B.; Wang, Y. Antibodies against a secreted protein from hookworm larvae reduce the intensity of hookworm infection in humans and vaccinated laboratory animals. FASEB J. 2005, 19, 1743–1745. [Google Scholar] [CrossRef]
- De Moira, A.P.; Fulford, A.J.C.; Kabatereine, N.B.; Ouma, J.H.; Booth, M.; Dunne, D.W. Analysis of complex patterns of human exposure and immunity to Schistosomiasis mansoni: The influence of age, sex, ethnicity and IgE. PLoS Negl. Trop. Dis. 2010, 4, e820. [Google Scholar] [CrossRef] [Green Version]
- Hirata, T.; Uchima, N.; Kishimoto, K.; Zaha, O.; Kinjo, N.; Hokama, A.; Sakugawa, H.; Kinjo, F.; Fujita, J. Impairment of host immune response against Strongyloides stercoralis by human T cell lymphotropic virus type 1 infection. Am. J. Trop. Med. Hyg. 2006, 74, 246–249. [Google Scholar] [CrossRef] [Green Version]
- Matowicka-karna, J.; Kemona, H. IgE antibodies in toxoplasmosis Przeciwciała IgE w toksoplazmozie. Postepy Hig. Med. Dosw. Online 2014, 68, 597–602. [Google Scholar] [CrossRef]
- Lindo, J.F.; Atkins, N.S.; Lee, M.G.; Robinson, R.D.; Bundy, D.A.P. long-term serum antibody isotype responses to Strongyloides stercoralis filariform antigens in eight patients treated with ivermectin. Am. J. Trop. Med. Hyg. 1996, 55, 474–476. [Google Scholar] [CrossRef]
- Rodrigues, R.M.; Sopelete, M.C.; De Oliveira Silva, D.A.; Cunha-Júnior, J.P.; Taketomi, E.A.; Costa-Cruz, J.M. Strongyloides ratti antigenic components recognized by IgE antibodies in immunoblotting as an additional tool for improving the immunodiagnosis in human strongyloidiasis. Mem. Inst. Oswaldo Cruz 2004, 99, 89–93. [Google Scholar] [CrossRef] [Green Version]
- De Jesus Inês, E.; Silva, M.L.S.; Souza, J.N.; Teixeira, M.C.A.; Soares, N.M. The role of glycosylated epitopes in the serodiagnosis of Strongyloides stercoralis infection. Diagn. Microbiol. Infect. Dis. 2013, 76, 31–35. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, H.; Balachandra, D.; Arifin, N.; Nolan, T.J.; Lok, J.B.; Hayat Khan, A.; Yunus, M.H.; Noordin, R. Diagnostic Potential of an IgE-ELISA in Detecting Strongyloidiasis. Am. J. Trop. Med. Hyg. 2020, 103, 2288–2293. [Google Scholar] [CrossRef]
- Yunus, M.H.; Arifin, N.; Balachandra, D.; Anuar, N.S.; Noordin, R. Lateral Flow Dipstick Test for Serodiagnosis of Strongyloidiasis. Am. J. Trop. Med. Hyg. 2019, 101, 432–435. [Google Scholar] [CrossRef]
- Arifin, N.; Yunus, M.H.; Nolan, T.J.; Lok, J.B.; Noordin, R. Identification and Preliminary Evaluation of a Novel Recombinant Protein for Serodiagnosis of Strongyloidiasis. Am. J. Trop. Med. Hyg. 2018, 98, 1165–1170. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2000. [Google Scholar]
- Krolewiecki, A.; Nutman, T.B. Strongyloidiasis: A Neglected Tropical Disease. Infect. Dis. Clin. N. Am. 2019, 33, 135–151. [Google Scholar] [CrossRef]
- Kennedy, M.B. Origin of Pdz (Dhr, Glgf) domains. Trends Biochem. Sci. 1995, 20, 350. [Google Scholar] [CrossRef]
- Harris, B.Z.; Lim, W.A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001, 114, 3219–3231. [Google Scholar] [CrossRef]
- Niethammer, M.; Kim, E.; Sheng, M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J. Neurosci. 1996, 16, 2157–2163. [Google Scholar] [CrossRef] [Green Version]
- Dev, K.K. Making protein interactions druggable: Targeting PDZ domains. Nat. Rev. Drug Discov. 2004, 3, 1047–1056. [Google Scholar] [CrossRef]
- Mu, Y.; Huang, H.; Liu, S.; Cai, P.; Gao, Y. Molecular characterization and ligand binding specificity of the PDZ domain-containing protein GIPC3 from Schistosoma japonicum. Parasit. Vectors 2012, 5, 227. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.M.; de Oliveira, M.C.; Sopelete, M.C.; Silva, D.A.O.; Campos, D.M.B.; Taketomi, E.A.; Costa-Cruz, J.M. IgG1, IgG4, and IgE antibody responses in human strongyloidiasis by ELISA using Strongyloides ratti saline extract as heterologous antigen. Parasitol. Res. 2007, 101, 1209–1214. [Google Scholar] [CrossRef]
- Varatharajalu, R.; Parandaman, V.; Ndao, M.; Andersen, J.F.; Neva, F.A. Strongyloides stercoralis excretory/secretory protein strongylastacin specifically recognized by IgE antibodies in infected human sera. Microbiol. Immunol. 2011, 55, 115–122. [Google Scholar] [CrossRef]
- Ramachandran, S.; Thompson, R.W.; Gam, A.A.; Neva, F.A. Recombinant cDNA clones for immunodiagnosis of strongyloidiasis. J. Infect. Dis. 1998, 177, 196–203. [Google Scholar] [CrossRef] [Green Version]

| Evaluation | IgE Detected Clones | |||||
|---|---|---|---|---|---|---|
| A132 | A133 | FB | A51 | A31 | 3A.1 | |
| Reactivity with positive serum (%), n = 10 | 100 | 100 | 87.5 | 100 | 100 | 100 |
| Non-reactivity with negative serum (%), n = 25 | 86 | 92 | 80 | 82 | 86 | 91 |
| Nucleotide Sequence Analysis | Amino Acid Sequence Analysis | |||||
|---|---|---|---|---|---|---|
| IgE Detective Clone | Gene Bank Accession Number | BLAST Result | Identity to Strongyloides sp. Sequence | Gene Bank Accession Number | BLAST Result | Identity to Strongyloides sp. Sequence |
| A132 | LL999048.1 | Strongyloides stercoralis genome assembly, S._stercoralis_PV0001, scaffold SSTP_scaffold0000001 | 100% | CEF67406.1 | Strongyloides ratti Oligosaccharyl transferase complex subunit (OSTC) | 96% |
| A133 | LL999051.1 | Strongyloides stercoralis genome assembly S._stercoralis_PV0001, scaffold SSTP_contig0000002 | 100% | CEF67580.1 | Strongyloides ratti PDZ signaling domain protein (GH21964p) | 87% |
| FB | LL999088.1 | Strongyloides stercoralis genome assembly, S._stercoralis_PV0001, scaffold SSTP_contig0000026 | 100% | CEF61968.1 | Strongyloides ratti 40S ribosomal protein S5 | 100% |
| A51 | LL999049.1 | Strongyloides stercoralis genome assembly S._stercoralis_PV0001, scaffold SSTP_scaffold0000002 | 100% | - | No significant similarities found | - |
| A31 | LL999050.1 | Strongyloides stercoralis genome assembly S._stercoralis_PV0001, scaffold SSTP_contig0000001 | 100% | CEF64458.1 | Strongyloides ratti, MSP domain and PapD-like domain-containing protein | 97% |
| 3A.1 | LL999051.1 | Strongyloides stercoralis genome assembly S._stercoralis_PV0001, scaffold SSTP_contig0000002 | 100% | CEF67580.1 | Strongyloides ratti PDZ signaling domain protein (GH21964p) | 84% |
| Samples | N | Reactivity | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Strongyloidiasis Group IA and Group IB | 43 | 43 | 0 | 100% | - |
| Group IA | 32 | 32 | 0 | - | - |
| Group IB | 11 | 11 | 0 | - | - |
| Negative Control Group II and Group III | 144 | 1 | 143 | 99.3% | |
| Other infections (Group 11) | - | ||||
| Amoebiasis | 4 | 0 | 4 | ||
| Ascariasis | 4 | 0 | 4 | - | - |
| Brµgian filariasis | 7 | 0 | 7 | - | - |
| Bancroftian filariasis | 4 | 0 | 4 | - | - |
| Hookworm infection | 8 | 1 | 8 | - | - |
| Hydatidosis | 4 | 0 | 4 | - | - |
| Schistosomiasis | 11 | 0 | 11 | - | - |
| Taeniasis | 2 | 0 | 2 | - | - |
| Toxocariasis | 19 | 0 | 19 | - | - |
| Toxoplasmosis | 5 | 0 | 5 | - | - |
| Trichuriasis | 2 | 0 | 2 | - | - |
| Giardiasis | 1 | 1 | |||
| Gnathostomiasis | 1 | 1 | |||
| Fascioliasis | 3 | 3 | |||
| Trichostrongylosis | 3 | 3 | |||
| Malaria | 5 | 5 | |||
| Onchocerciasis | 2 | 2 | |||
| Loaisis | 1 | 1 | |||
| Mixed infection (ascaris/trichuris/hookworm) | 3 | 0 | 3 | ||
| Healthy donors (Group III) | 55 | 0 | 55 | - | - |
| Serum samples from patients with high IgE due to allergies (excluded from diagnostic specificity determination) | 4 | 0 | 4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, H.; Arifin, N.; Nolan, T.J.; Lok, J.B.; Anuar, N.S.; Noordin, R. Strongyloides-Specific IgE Phage cDNA Clones and Development of a Novel ELISA for Strongyloidiasis. Diagnostics 2021, 11, 985. https://doi.org/10.3390/diagnostics11060985
Ahmad H, Arifin N, Nolan TJ, Lok JB, Anuar NS, Noordin R. Strongyloides-Specific IgE Phage cDNA Clones and Development of a Novel ELISA for Strongyloidiasis. Diagnostics. 2021; 11(6):985. https://doi.org/10.3390/diagnostics11060985
Chicago/Turabian StyleAhmad, Hussain, Norsyahida Arifin, Thomas J. Nolan, James B. Lok, Nor Suhada Anuar, and Rahmah Noordin. 2021. "Strongyloides-Specific IgE Phage cDNA Clones and Development of a Novel ELISA for Strongyloidiasis" Diagnostics 11, no. 6: 985. https://doi.org/10.3390/diagnostics11060985






