Legionella pneumophila—Epidemiology and Characterization of Clinical Isolates, Slovenia, 2006–2020
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Routine Legionella Testing
2.2. Urine Antigen Test (UAT)
2.3. Polymerase Chain Reaction (PCR)
2.4. Legionella spp. Cultivation
2.5. Phenotypic and Genotypic Analysis
2.6. Serology
3. Results
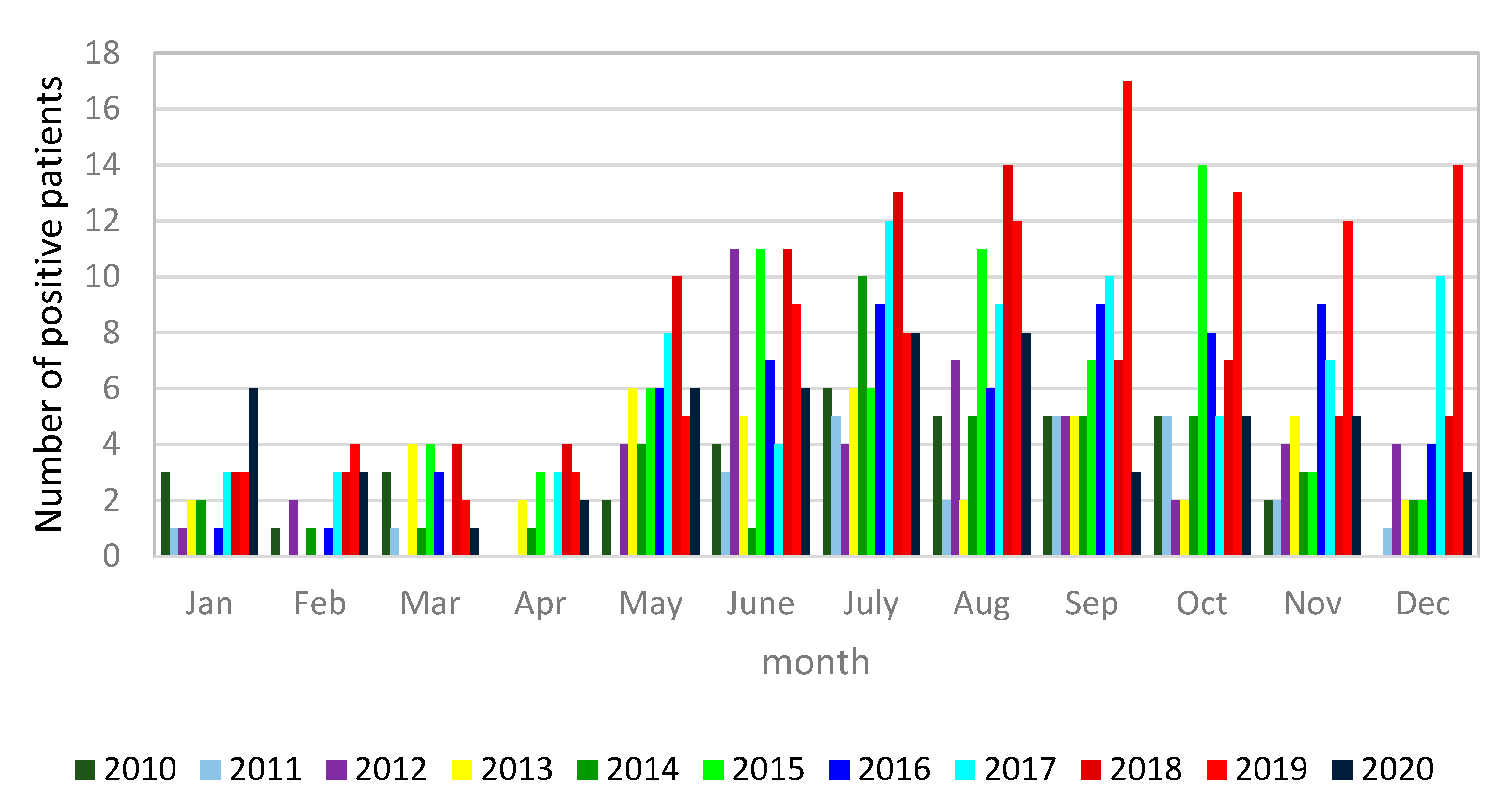
3.1. Microbiological Diagnostics of Legionella pneumophila Infections
3.2. Legionella pneumophila Phenotyping and Genotyping Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guyard, C.; Low, D.E. Legionella infections and travel associated legionellosis. Travel Med. Infect. Dis. 2011, 9, 176–186. [Google Scholar] [CrossRef]
- Buchrieser, C. Legionella: From protozoa to humans. Front. Microbiol. 2011, 2, 182. [Google Scholar] [CrossRef] [Green Version]
- Carratalà, J.; Garcia-Vidal, C. An update on Legionella. Curr. Opin. Infect. Dis. 2010, 23, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Shames, S.R. Pathogenicity and Virulence ofLegionella: Intracellular replication and host response. Virulence 2021, 12, 1122–1144. [Google Scholar] [CrossRef]
- Dagan, A.; Epstein, D.; Mahagneh, A.; Nashashibi, J.; Geffen, Y.; Neuberger, A.; Miller, A. Community-acquired versus nosocomial Legionella pneumonia: Factors associated with Legionella-related mortality. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Diederen, B. Legionella spp. and Legionnaires’ disease. J. Infect. 2008, 56, 1–12. [Google Scholar] [CrossRef]
- Commission Implementing Decision (EU) 2018/945 of 22 June 2018 on the Com-municable Diseases and Related Special Health Issues to Be Covered by Epidemi-ological Surveillance as Well as Relevant Case Definitions (Text with EEA Rele-vance.). 2018, Volume 170. Available online: https://eur-lex.europa.eu/eli/dec_impl/2018/945/oj (accessed on 30 June 2021).
- Beauté, J.; Robesyn, E.; De Jong, B. Legionnaires’ disease in Europe: All quiet on the eastern front? Eur. Respir. J. 2013, 42, 1454–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, F.B.; Schmutz, C.; Gaia, V.; Mäusezahl, D. Legionnaires’ Disease on the Rise in Switzerland: A Denominator-Based Analysis of National Diagnostic Data, 2007–2016. Int. J. Environ. Res. Public Health 2020, 17, 7343. [Google Scholar] [CrossRef]
- Beauté, J. On behalf of the European Legionnaires’ Disease Surveillance Network Legionnaires’ Disease in Europe, 2011 to 2015. Eurosurveillance 2017, 22. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control. Annual Epidemiological Report for 2017. Legionnaires’ Disease; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2019. [Google Scholar]
- SiStat Database. Available online: https://pxweb.stat.si/sistat/en (accessed on 15 June 2021).
- Mentasti, M.; Kese, D.; Echahidi, F.; Uldum, S.A.; Afshar, B.; David, S.; Mrazek, J.; De Mendonça, R.; Harrison, T.G.; Chalker, V.J. Design and validation of a qPCR assay for accurate detection and initial serogrouping of Legionella pneumophila in clinical specimens by the ESCMID Study Group for Legionella Infections (ESGLI). Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1387–1393. [Google Scholar] [CrossRef]
- Harrison, T.G.; Taylor, A.G. A Laboratory Manual for Legionella; John Wiley and Sons: Chichester, UK, 1988; pp. 13–30. [Google Scholar]
- Lück, C.; Fry, N.K.; Helbig, J.H.; Jarraud, S.; Harrison, T.G. Typing Methods for Le-gionella. In Legionella: Methods and Protocols; Buchrieser, C., Hilbi, H., Eds.; Hu-mana Press: Totowa, NJ, USA, 2013; pp. 119–148. ISBN 978-1-62703-161-5. [Google Scholar]
- Gaia, V.; Fry, N.K.; Afshar, B.; Luück, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus Sequence-Based Scheme for Epidemiological Typing of Clinical and Environmental Isolates of Legionella pneumophila. J. Clin. Microbiol. 2005, 43, 2047–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratzow, S.; Gaia, V.; Helbig, J.H.; Fry, N.; Lück, P.C. Addition of neuA, the Gene Encoding N-Acylneuraminate Cytidylyl Transferase, Increases the Discriminatory Ability of the Consensus Sequence-Based Scheme for Typing Legionella pneumophila Serogroup 1 Strains. J. Clin. Microbiol. 2007, 45, 1965–1968. [Google Scholar] [CrossRef] [Green Version]
- Ginevra, C.; Lopez, M.; Forey, F.; Reyrolle, M.; Meugnier, H.; Vandenesch, F.; Etienne, J.; Jarraud, S.; Molmeret, M. Evaluation of a Nested-PCR-Derived Sequence-Based Typing Method Applied Directly to Respiratory Samples from Patients with Legionnaires’ Disease. J. Clin. Microbiol. 2009, 47, 981–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarraud, S.; Descours, G.; Ginevra, C.; Lina, G.; Etienne, J. Identification of Legionella in Clinical Samples. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 954, pp. 27–56. [Google Scholar]
- Campèse, C.; Descours, G.; Lepoutre, A.; Beraud, L.; Maine, C.; Che, D.; Jarraud, S. Legionnaires’ disease in France. Médecine Mal. Infect. 2015, 45, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Phin, N.; Parry-Ford, F.; Harrison, T.; Stagg, H.; Zhang, N.; Kumar, K.; Lortholary, O.; Zumla, A.; Abubakar, I. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect. Dis. 2014, 14, 1011–1021. [Google Scholar] [CrossRef]
- Rota, M.C.; Caporali, M.G.; Bella, A.; Ricci, M.L.; Napoli, C. Legionnaires’ disease in Italy: Results of the epidemiological surveillance from 2000 to 2011. Eurosurveillance 2013, 18, 20497. [Google Scholar] [CrossRef] [Green Version]
- Toberna, C.P.; William, H.M.; Kram, J.J.F.; Heslin, K.; Baumgardner, D.J. Epidemiologic Survey of Legionella Urine Antigen Testing Within a Large Wisconsin-Based Health Care System. J. Patient-Centered Res. Rev. 2020, 7, 165–175. [Google Scholar] [CrossRef]
- Muyldermans, A.; On behalf of the National Expert Committee on Infectious Serology; Descheemaeker, P.; Boel, A.; Desmet, S.; Van Gasse, N.; Reynders, M. What is the risk of missing legionellosis relying on urinary antigen testing solely? A retrospective Belgian multicenter study. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 39, 729–734. [Google Scholar] [CrossRef]
- Cunha, B.; Burillo, A.; Bouza, E. Legionnaires’ disease. Lancet 2016, 387, 376–385. [Google Scholar] [CrossRef]
- Elverdal, P.; Jørgensen, C.; Krogfelt, K.A.; Uldum, S. Two years’ performance of an in-house ELISA for diagnosis of Legionnaires’ disease: Detection of specific IgM and IgG antibodies against Legionella pneumophila serogroup 1, 3 and 6 in human serum. J. Microbiol. Methods 2013, 94, 94–97. [Google Scholar] [CrossRef]
- Peci, A.; Winter, A.-L.; Gubbay, J.B. Evaluation and Comparison of Multiple Test Methods, Including Real-time PCR, for Legionella Detection in Clinical Specimens. Front. Public Health 2016, 4, 175. [Google Scholar] [CrossRef]
- Skaza, A.T.; Beskovnik, L.; Storman, A.; Kese, D.; Ursic, S. Epidemiological Inves-tigation of a Legionellosis Outbreak in a Slovenian Nursing Home, August 2010. Scand. J. Infect. Dis. 2012, 44, 263–269. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Annual Epidemiological Report for 2015. Legionnaires’ Disease; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2017. [Google Scholar]
- Sočan, M.; Frelih, T.; Klavs, I.; Grilc, E.; Grgič Vitek, M.; Učakar, V. Epidemiološko Spremljanje Nalezljivih Bolezni v Sloveniji v Letu 2018; Nacionalni inštitut za javno zdravje: Ljubljana, Slovenia, 2019. [Google Scholar]
- Fontana, S.; Scaturro, M.; Rota, M.C.; Caporali, M.G.; Ricci, M.L. Molecular Typing of Legionella Pneumophila Serogroup 1 Clinical Strains Isolated in Italy. Int. J. Med. Microbiol. 2014, 304, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Busó, L.; Coscollà, M.; Palero, F.; Camaró, M.L.; Gimeno, A.; Moreno, P.; Escribano, I.; López Perezagua, M.M.; Colomina, J.; Vanaclocha, H.; et al. Geographical and Temporal Structures of Legionella Pneumoph-ila Sequence Types in Comunitat Valenciana (Spain), 1998 to 2013. Appl. Environ. Microbiol. 2015, 81, 7106–7113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lévesque, S.; Lalancette, C.; Bernard, K.; Pacheco, A.L.; Dion, R.; Longtin, J.; Trem-blay, C. Molecular Typing of Legionella Pneumophila Isolates in the Province of Quebec from 2005 to 2015. PLoS ONE 2016, 11, e0163818. [Google Scholar] [CrossRef]
- Amemura-Maekawa, J.; Kura, F.; Chida, K.; Ohya, H.; Kanatani, J.; Isobe, J.; Tanaka, S.; Nakajima, H.; Hiratsuka, T.; Yoshino, S.; et al. Legionella Pneumophila and Other Legionella Species Isolated from Legionellosis Patients in Japan between 2008 and 2016. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginevra, C.; Chastang, J.; David, S.; Mentasti, M.; Yakunin, E.; Chalker, V.; Chalifa-Caspi, V.; Valinsky, L.; Jarraud, S.; Moran-Gilad, J.; et al. A real-time PCR for specific detection of the Legionella pneumophila serogroup 1 ST1 complex. Clin. Microbiol. Infect. 2020, 26, 514.e1–514.e6. [Google Scholar] [CrossRef]
- David, S.; Rusniok, C.; Mentasti, M.; Gomez-Valero, L.; Harris, S.R.; Lechat, P.; Lees, J.; Ginevra, C.; Glaser, P.; Ma, L.; et al. Multiple major disease-associated clones of Legionella pneumophila have emerged recently and independently. Genome Res. 2016, 26, 1555–1564. [Google Scholar] [CrossRef] [Green Version]





| L. pneumophila Serogroup (n/%) | Phenotype (n/%) | Sequence Types (n) |
|---|---|---|
| L. pneumophila sg 1 (82/93.2%) | Allentown/France (36/43.9%) | ST23 (15); ST82 (4); ST1852 (4); ST62 (3); ST203 (3); ST762 (2); ST1983 (2); ST1 (1); ST109 (1); ST224 (1); |
| Philadelphia (24/29.3%) | ST1 (14); ST37 (4); ST862 (2); ST62 (1), ST146 (1); ST435 (1); ST1090 (1) | |
| Bellingham (9/11.0%) | ST728 (4); ST334 (2); ST59 (1); ST196 (1); ST763 (1) | |
| Knoxville (7/8.5%) | ST1 (1); ST9 (1); ST20 (2); ST23 (2); ST62 (1) | |
| Benidorm (4/4.9%) | ST1299 (2); ST477 (1); ST1984 (1) | |
| Oxford/Olda (2/2.4%) | ST1 (2) | |
| L. pneumophila sg 2-14 (6/6.8%) | Bloomington 1 (1/16.7%) | ST93 (1) |
| Chicago 2 (2/33.3%) | ST421 (1); ST2999 (1) | |
| Concord 3 (1/16.7%) | ST1324 (1) | |
| 570 CO H (1/16.7%) | ST421 (1) | |
| ND (1/16.7%) | ST2998 (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keše, D.; Obreza, A.; Rojko, T.; Kišek, T.C. Legionella pneumophila—Epidemiology and Characterization of Clinical Isolates, Slovenia, 2006–2020. Diagnostics 2021, 11, 1201. https://doi.org/10.3390/diagnostics11071201
Keše D, Obreza A, Rojko T, Kišek TC. Legionella pneumophila—Epidemiology and Characterization of Clinical Isolates, Slovenia, 2006–2020. Diagnostics. 2021; 11(7):1201. https://doi.org/10.3390/diagnostics11071201
Chicago/Turabian StyleKeše, Darja, Aljoša Obreza, Tereza Rojko, and Tjaša Cerar Kišek. 2021. "Legionella pneumophila—Epidemiology and Characterization of Clinical Isolates, Slovenia, 2006–2020" Diagnostics 11, no. 7: 1201. https://doi.org/10.3390/diagnostics11071201






