Differentiating Breast Tumors from Background Parenchymal Enhancement at Contrast-Enhanced Mammography: The Role of Radiomics—A Pilot Reader Study
Abstract
:1. Introduction
2. Materials and Methods
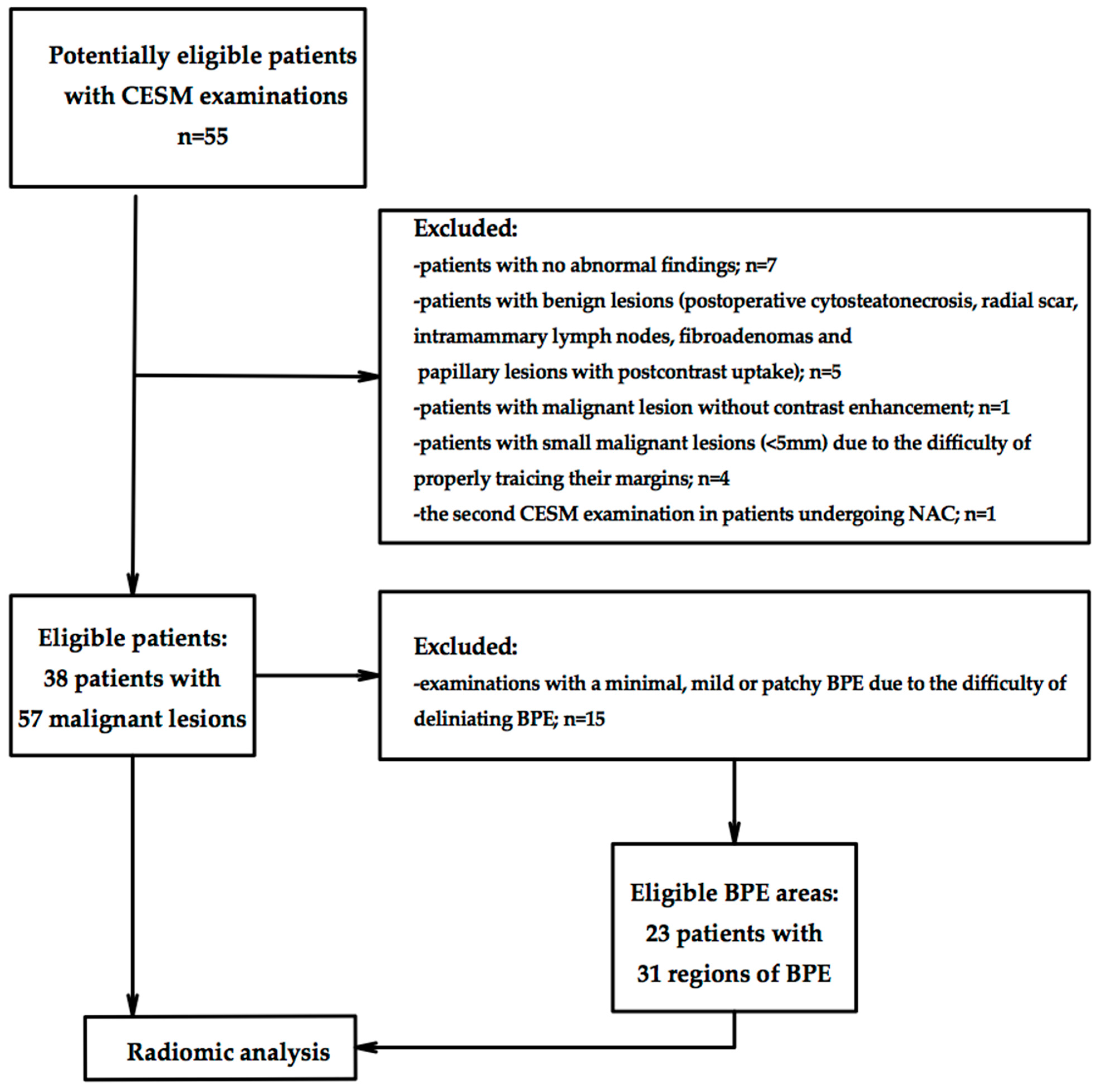
2.1. Study Population
2.2. Image Acquisition and Interpretation
2.3. Reference Standard
2.4. Texture Analysis Protocol
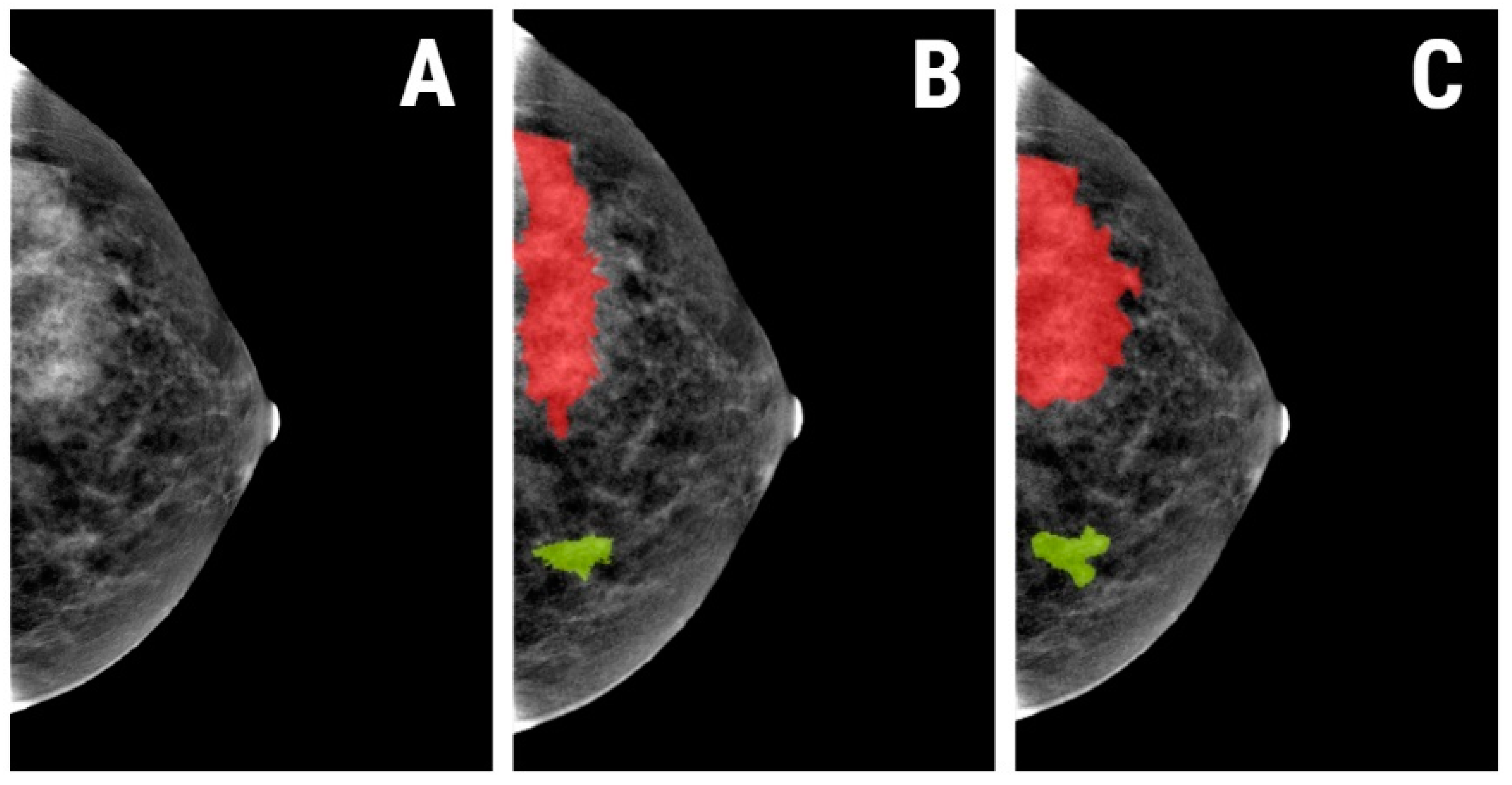
2.4.1. Image Pre-Processing and Segmentation
2.4.2. Feature Extraction
2.4.3. Feature Selection
2.4.4. Class Prediction
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Fallenberg, E.M.; Dromain, C.; Diekmann, F.; Engelken, F.; Krohn, M.; Singh, J.M.; Ingold-Heppner, B.; Winzer, K.J.; Bick, U.; Renz, A.D. Contrast-enhanced spectral mammography versus MRI: Initial results in the detection of breast cancer and assessment of tumour size. Eur. Radiol. 2014, 24, 256–264. [Google Scholar] [CrossRef]
- Thibault, F.; Balleyguier, C.; Tardivon, A.; Dromain, C. Contrast enhanced spectral mammography: Better than MRI? Eur. J. Radiol. 2012, 81 (Suppl. S1), S162–S164. [Google Scholar] [CrossRef]
- Jochelson, M.S.; Dershaw, D.D.; Sung, J.S.; Heerdt, A.S.; Thornton, C.; Moskowitz, C.S.; Ferrara, J.; Morris, E.A. Bilateral contrast-enhanced dual-energy digital mammography: Feasibility and comparison with conventional digital mammography and MR imaging in women with known breast carcinoma. Radiology 2013, 266, 743–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbs, M.M.; Taylor, D.B.; Buzynski, S.; Peake, R.E. Contrast-enhanced spectral mammography (CESM) and contrast enhanced MRI (CEMRI): Patient preferences and tolerance. J. Med. Imaging Radiat. Oncol. 2015, 59, 300–305. [Google Scholar] [CrossRef]
- Lobbes, M.B.I.; Lalji, U.; Houwers, J.; Nijssen, E.C.; Nelemans, P.J.; Van Roozendaal, L.; Smidt, M.L.; Heuts, E.; Wildberger, J.E. Contrast-enhanced spectral mammography in patients referred from the breast cancer screening programme. Eur. Radiol. 2014, 24, 1668–1676. [Google Scholar] [CrossRef] [PubMed]
- ElSaid, N.A.E.S.; Mahmoud, H.G.M.; Salama, A.; Nabil, M.; ElDesouky, E.D. Role of contrast enhanced spectral mammography in predicting pathological response of locally advanced breast cancer post neo-adjuvant chemotherapy. Egypt J. Radiol. Nucl. Med. 2017, 48, 519–527. [Google Scholar] [CrossRef]
- Losurdo, L.; Fanizzi, A.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Dentamaro, R.; Didonna, V.; Lorusso, V.; Massafra, R.; Tamborra, P.; et al. Radiomics analysis on contrast-enhanced spectral mammography images for breast cancer diagnosis: A pilot study. Entropy 2019, 21, 1110. [Google Scholar] [CrossRef] [Green Version]
- Sogani, J.; Morris, E.A.; Kaplan, J.B.; D’Alessio, D.; Goldman, D.; Moskowitz, C.S.; Jochelson, M.S. Comparison of background parenchymal enhancement at contrast-enhanced spectral mammography and breast MR imaging. Radiology 2017, 282, 63–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uematsu, T.; Kasami, M.; Watanabe, J. Does the degree of background enhancement in breast MRI affect the detection and staging of breast cancer? Eur. Radiol. 2011, 21, 2261–2267. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Gastounioti, A.; Conant, E.F.; Kontos, D. Beyond breast density: A review on the advancing role of parenchymal texture analysis in breast cancer risk assessment. Breast Cancer Res. 2016, 18, 91. [Google Scholar] [CrossRef] [Green Version]
- Collewet, G.; Strzelecki, M.; Mariette, F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging 2004, 22, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Roy, A.K. Wavelet Based Texture Analysis for Medical Images. Int. J. Adv. Res. Electr. Electron. Instrum. Eng. 2015, 4, 3958–3963. [Google Scholar]
- Livens, S.; Scheunders, P.; Van de Wouwer, G.; Van Dyck, D. Wavelets for texture analysis, an overview. In Proceedings of the 6th International Conference on Image Processing and its Applications, Dublin, Ireland, 14–17 July 1997; pp. 581–585. [Google Scholar]
- Wang, Y.M.; Fan, W.; Zhang, K.; Zhang, L.; Tan, Z.; Ma, R. Comparison of transducers with different frequencies in breast contrast-enhanced ultrasound (CEUS) using SonoVue as contrast agent. Br. J. Radiol. 2016, 89, 20151050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, M.A.; Leithner, D.; Sung, J.; Avendano, D.; Morris, E.A.; Pinker, K.; Jochelson, M.S. Radiomics for tumor characterization in breast cancer patients: A feasibility study comparing contrast-enhanced mammography and magnetic resonance imaging. Diagnostics 2020, 10, 492. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.A.; Pinker, K.; Leithner, D.; Sung, J.; Avendano, D.; Morris, E.A.; Jochelson, M. Contrast-Enhanced Mammography and Radiomics Analysis for Noninvasive Breast Cancer Characterization: Initial Results. Mol. Imaging Biol. 2020, 22, 780–787. [Google Scholar] [CrossRef]
- la Forgia, D.; Fanizzi, A.; Campobasso, F.; Bellotti, R.; Didonna, V.; Lorusso, V.; Moschetta, M.; Massafra, R.; Tamborra, P.; Tangaro, S.; et al. Radiomic analysis in contrast-enhanced spectral mammography for predicting breast cancer histological outcome. Diagnostics 2020, 10, 708. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, F.; Ma, H.; Shi, Y.; Dong, J.; Yang, P.; Zhang, K.; Guo, N.; Zhang, R.; Cui, J.; et al. Contrast-Enhanced Spectral Mammography-Based Radiomics Nomogram for the Prediction of Neoadjuvant Chemotherapy-Insensitive Breast Cancers. Front. Oncol. 2021, 11, 84. [Google Scholar]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Plecha, D.; Madabhushi, A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017, 19, 57. [Google Scholar] [CrossRef]
- Fanizzi, A.; Losurdo, L.; Basile, T.M.A.; Bellotti, R.; Bottigli, U.; Delogu, P.; Diacono, D.; Didonna, V.; Fausto, A.; Lombardi, A.; et al. Fully Automated Support System for Diagnosis of Breast Cancer in Contrast-Enhanced Spectral Mammography Images. J. Clin. Med. 2019, 8, 891. [Google Scholar] [CrossRef] [Green Version]
- Losurdo, L.; Basile, T.M.A.; Fanizzi, A.; Bellotti, R.; Bottigli, U.; Carbonara, R.; Dentamaro, R.; Diacono, D.; Didonna, V.; Lombardi, A.; et al. A Gradient-Based Approach for Breast DCE-MRI Analysis. Biomed. Res. Int. 2018, 2018, 9032408. [Google Scholar] [CrossRef]
- Wang, H.; Hu, Y.; Li, H.; Xie, Y.; Wang, X.; Wan, W. Preliminary study on identification of estrogen receptor-positive breast cancer subtypes based on dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) texture analysis. Gland Surg. 2020, 9, 622–628. [Google Scholar] [CrossRef]
- Tagliafico, A.; Bignotti, B.; Tagliafico, G.; Tosto, S.; Signori, A.; Calabrese, M. Quantitative evaluation of background parenchymal enhancement (BPE) on breast MRI. A feasibility study with a semi-automatic and automatic software compared to observer-based scores. Br. J. Radiol. 2015, 88, 20150417. [Google Scholar] [CrossRef] [Green Version]
- Sutton, E.J.; Huang, E.P.; Drukker, K.; Burnside, E.S.; Li, H.; Net, J.M.; Rao, A.; Whitman, G.J.; Zuley, M.; Ganott, M.; et al. Breast MRI radiomics: Comparison of computer- and human-extracted imaging phenotypes. Eur. Radiol. Exp. 2017, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Mazurowski, M.A.; Zhang, J.; Grimm, L.J.; Yoon, S.C.; Silber, J.I. Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014, 273, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kato, F.; Oyama-Manabe, N.; Li, R.; Cui, Y.; Tha, K.K.; Yamashita, H.; Kudo, K.; Shirato, H. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: A pilot radiomics study. PLoS ONE 2015, 10, e0143308. [Google Scholar] [CrossRef]
- Wu, S.; Berg, W.A.; Zuley, M.L.; Kurland, B.F.; Jankowitz, R.C.; Nishikawa, R.; Gur, D.; Sumkin, J.H. Breast MRI contrast enhancement kinetics of normal parenchyma correlate with presence of breast cancer. Breast Cancer Res. 2016, 18, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilorenzo, G.; Telegrafo, M.; La Forgia, D.; Stabile Ianora, A.A.; Moschetta, M. Breast MRI background parenchymal enhancement as an imaging bridge to molecular cancer sub-type. Eur. J. Radiol. 2019, 113, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, B.; Sun, X.; Cao, G.; Rubin, D.L.; Napel, S.; Ikeda, D.M.; Kurian, A.W.; Li, R. Heterogeneous enhancement patterns of tumor-adjacent parenchyma at MR imaging are associated with dysregulated signaling pathways and poor survival in breast cancer. Radiology 2017, 285, 401–413. [Google Scholar] [CrossRef] [Green Version]
| Class | Parameters | Number of Parameters | Variations | Computation |
|---|---|---|---|---|
| Absolute gradient | GrMean, GrVariance, GrSkewness, GrKurtosis, GrNonZeros, and percentage of pixels with nonzero gradient | 5 | - | 4 bits/pixel |
| Histogram | Mean, Variance, Skewness, Kurtosis, and Perc.01–99% | 5 | - | - |
| Run Length Matrix | RLNonUni, GLevNonU, LngREmph, ShrtREmp, and Fraction | 20 | 4 directions | 6 bits/pixel |
| Co-occurrence Matrix | AngScMom, Contrast, Correlat, SumOfSqs, InvDfMom, SumAverg, SumVarnc, SumEntrp, Entropy, DifVarnc, and DifEntrp | 220 | 4 directions | 6 bits/pixel; 5 between-pixels distances |
| Auto-regressive Model | Teta 1–4 andSigma | 5 | - | - |
| Wavelet transformation | WavEn | 20 | 4 frequency bands | 5 scales |
| Age | Pathological Results | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nottingham | Hormonal Status | Her2 | Ki67 | |||||||
| I | II | III | ER+ vs. ER− | PR+ vs. PR− | + | − | <14% | ≥14% | ||
| Training group | 30–71 (mean 44) | 11/57 (19.2%) | 26/57 (46.6%) | 20/57 (35.1%) | 51/57 (89.4%) vs. 6/57 (10.5%) | 45/57 (78.9%) vs. 12/57 (21%) | 9/57 (15.7%) | 48/57 (84.2%) | 15/57 (26.3%) | 42/57 (73.6%) |
| Validating group | 40–68 (mean 49.4) | 1/5 (20%) | 3/5 (60%) | 1/5 (20%) | 4/5 (80%) vs. 1/5 (20%) | 2/5 (40%) vs. 3/5 (60%) | 1/5 (20%) | 4/5 (80%) | 1/5 (20%) | 4/5 (80%) |
| Parameter | p-Value M-W | Malignant Lesions | BPE | ||
|---|---|---|---|---|---|
| Median | IQR | Median | IQR | ||
| Fisher | |||||
| CZ5D6SumAverg | <0.0001 | 53.82 | 43.43–68.16 | 41.74 | 36.56–45.22 |
| CN5D6SumAverg | <0.0001 | 53.72 | 43.5–68.12 | 41.85 | 36.59–45.14 |
| CZ4D6SumAverg | <0.0001 | 53.68 | 43.38–68.08 | 41.7 | 36.53–45.16 |
| Perc10 | <0.0001 | 75 | 59.5–94.75 | 58 | 48.5–63 |
| CN4D6SumAverg | <0.0001 | 53.6 | 43.43–68.04 | 41.79 | 36.55–45.09 |
| CV5D6SumAverg | <0.0001 | 53.55 | 43.39–68.03 | 41.75 | 36.5–45.09 |
| CH5D6SumAverg | <0.0001 | 53.61 | 43.36–68.04 | 41.67 | 36.53–45.17 |
| CZ3D6SumAverg | <0.0001 | 53.49 | 43.31–67.98 | 41.66 | 36.49–45.09 |
| CV4D6SumAverg | <0.0001 | 53.44 | 43.34–67.96 | 41.71 | 36.48–45.04 |
| CN3D6SumAverg | <0.0001 | 53.46 | 43.36–67.96 | 41.73 | 36.51–45.02 |
| POE + ACC | |||||
| Kurtosis | 0.0007 | −0.1448 | −0.3243 to −0.02027 | −0.003272 | −0.06681 to 0.1240 |
| RHD6ShrtREmp | 0.1916 | 0.915 | 0.9092 to 0.9195 | 0.9114 | 0.9089 to 0.9179 |
| Skewness | 0.0007 | 0.006708 | −0.1117 to 0.1364 | 0.09351 | 0.03894 to 0.2256 |
| CZ5D6Correlat | 0.0047 | 0.5239 | 0.3777 to 0.7047 | 0.4212 | 0.2712 to 0.5196 |
| ATeta3 | 0.0563 | 0.3143 | 0.3075 to 0.3308 | 0.3081 | 0.3000 to 0.3212 |
| CN4D6Correlat | 0.0009 | 0.5855 | 0.3871 to 0.7316 | 0.4671 | 0.2817 to 0.5469 |
| GD4Kurtosis | 0.3432 | 0.295 | 0.2392 to 0.3738 | 0.2973 | 0.2242 to 0.3229 |
| Perc01 | 0.0001 | 51 | 43.75–67 | 40 | 30.5000 to 47.0000 |
| RVD6ShrtREmp | 0.5851 | 0.9151 | 0.9105 to 0.9199 | 0.9138 | 0.9077 to 0.9192 |
| CH1D6SumAverg | <0.0001 | 53.08 | 43.16–67.73 | 41.57 | 36.4–44.84 |
| Mutual Information | |||||
| CZ2D6DifEntrp | 0.0829 | 1.02 | 1–1.04 | 0.9988 | 0.98–1.03 |
| WavEnLL_s-1 | <0.0001 | 11,307.18 | 7250.37–18,682.45 | 6814.86 | 5219.29–7871.86 |
| WavEnLL_s-2 | <0.0001 | 11,213.75 | 7112.81–18,445.7 | 6724.37 | 5125.37–7693.77 |
| CH2D6SumAverg | <0.0001 | 53.17 | 43.21–67.81 | 41.59 | 36.43–44.94 |
| CH3D6SumAverg | <0.0001 | 53.32 | 43.27–67.89 | 41.62 | 36.47–45.03 |
| CH4D6SumAverg | <0.0001 | 53.46 | 43.32–67.97 | 41.64 | 36.5–45.1 |
| CN1D6SumAverg | <0.0001 | 53.09 | 43.19–67.77 | 41.61 | 36.41–44.86 |
| Parameter | Coefficient | Standard Error | p-Value | VIF |
|---|---|---|---|---|
| CH1D6SumAverg | 0.017 | 0.041 | 0.67 | 250.56 |
| CN4D6Correlat | 0.308 | 0.552 | 0.578 | 5.887 |
| Kurtosis | −0.307 | 0.2337 | 0.192 | 2.565 |
| Perc01 | −0.018 | 0.021 | 0.387 | 71.517 |
| Perc10 | 0.03 | 0.035 | 0.3847 | 364.574 |
| Skewness | −0.366 | 0.386 | 0.346 | 4.708 |
| WavEnLL_s_2 | <−0.001 | <0.001 | 0.035 | 49.354 |
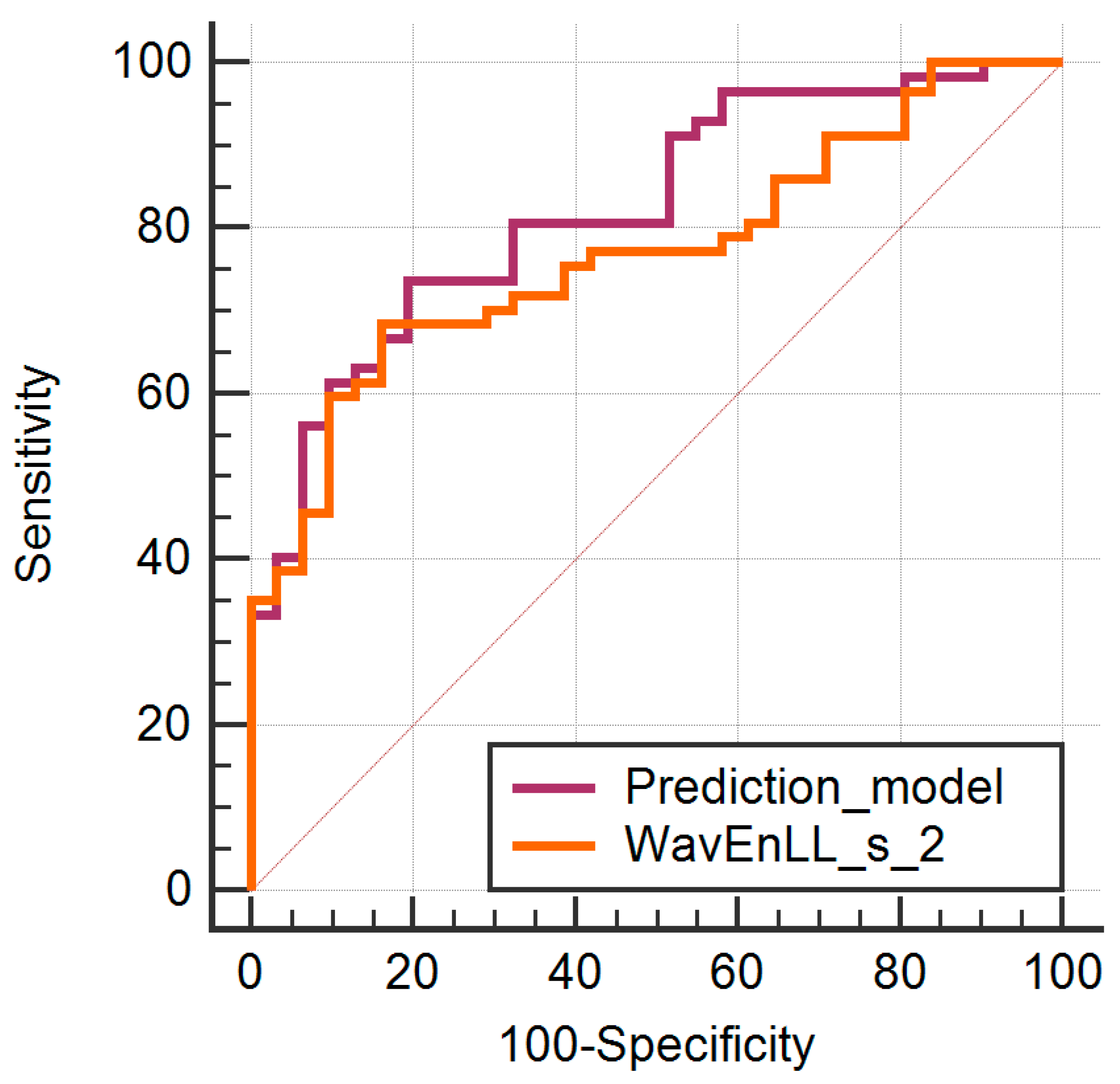
| Parameter | AUC | Sign.lvl. | J | Cut-Off | Se (%) | Sp (%) |
|---|---|---|---|---|---|---|
| WavEnLL_s_2 | 0.771 (0.67–0.854) | <0.0001 | 0.5229 | >8082.88 | 68.42 (54.8–80.1) | 83.87 (66.3–94.5) |
| Prediction model | 0.824 (0.728–0.897) | <0.0001 | 0.5433 | >0.55 | 73.68 (60.3–84.5) | 80.65 (62.5–92.5) |
| Misclassified Lesions | Accuracy (%) | Se (%) | Sp (%) | |||
|---|---|---|---|---|---|---|
| Total | Cancer | BPE | ||||
| Training group | 33/88 (37.5%) | 16/57 (28%) | 17/31 (54.8%) | 62.50 (51.53–72.6) | 71.93 (58.46–83.03) | 45.16 (27.32–63.97) |
| Validation group | 8/19 (42.1%) | 4/10 (40%) | 5/9 (55.5%) | 52.63 (28.86–75.55) | 60 (26.24–87.84) | 44.44 (13.70–78.8) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boca, I.; Ciurea, A.I.; Ciortea, C.A.; Ștefan, P.A.; Lisencu, L.A.; Dudea, S.M. Differentiating Breast Tumors from Background Parenchymal Enhancement at Contrast-Enhanced Mammography: The Role of Radiomics—A Pilot Reader Study. Diagnostics 2021, 11, 1248. https://doi.org/10.3390/diagnostics11071248
Boca I, Ciurea AI, Ciortea CA, Ștefan PA, Lisencu LA, Dudea SM. Differentiating Breast Tumors from Background Parenchymal Enhancement at Contrast-Enhanced Mammography: The Role of Radiomics—A Pilot Reader Study. Diagnostics. 2021; 11(7):1248. https://doi.org/10.3390/diagnostics11071248
Chicago/Turabian StyleBoca (Bene), Ioana, Anca Ileana Ciurea, Cristiana Augusta Ciortea, Paul Andrei Ștefan, Lorena Alexandra Lisencu, and Sorin Marian Dudea. 2021. "Differentiating Breast Tumors from Background Parenchymal Enhancement at Contrast-Enhanced Mammography: The Role of Radiomics—A Pilot Reader Study" Diagnostics 11, no. 7: 1248. https://doi.org/10.3390/diagnostics11071248
APA StyleBoca, I., Ciurea, A. I., Ciortea, C. A., Ștefan, P. A., Lisencu, L. A., & Dudea, S. M. (2021). Differentiating Breast Tumors from Background Parenchymal Enhancement at Contrast-Enhanced Mammography: The Role of Radiomics—A Pilot Reader Study. Diagnostics, 11(7), 1248. https://doi.org/10.3390/diagnostics11071248








