Muscle Synergy of Lower Limb Motion in Subjects with and without Knee Pathology
Abstract
:1. Introduction
2. Algorithm for Muscle Synergy Analysis
2.1. Muscle Synergy Extraction
2.2. Quantify the Similarity between Muscle Synergies
3. Materials and Methods
3.1. Data Description
3.2. Data Preprocessing
3.3. Data Analysis
3.3.1. Non-Negative Matrix Factorization (NMF) Decomposition for Each Segment
3.3.2. Building Representative Muscle Synergy for Each Motion of Each Subject
3.3.3. Similarity Analysis
3.4. NMF-Based Classification of Lower Limb Motions with sEMG Data
3.4.1. Segmentation
3.4.2. Feature Extraction
- RMS.
- Fourth-order auto-regressive model (AR).
- Interquartile Range (IQR).
- Waveform Length (WL).
- Mean Absolute Value (MAV).
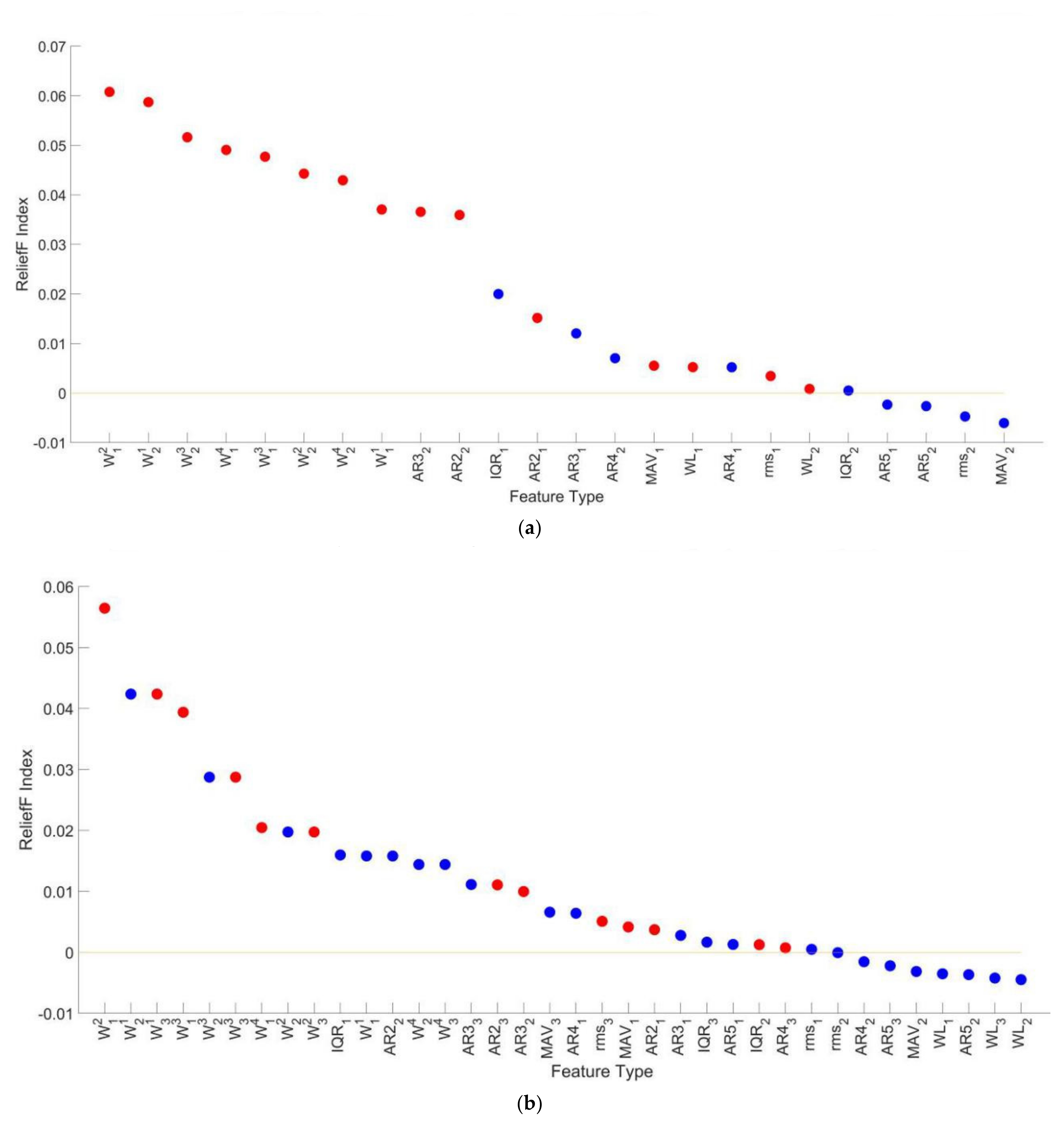
3.4.3. Feature Selection
3.4.4. Data Classification and Cross Validation
3.5. Diagnosis of Knee Pathology with NMF of sEMG Data
3.6. Statistical Analysis
4. Results
4.1. Optimal Number of Muscle Synergies
4.2. Muscle Synergy Similarities
4.3. Lower Limb Motions Classification
4.4. Knee Pathology Identification
5. Discussion
5.1. Muscle Synergy Differences between Different Subject Groups or Different Motions
5.2. Performance of Muscle Synergy-Based Lower Limb Motion Classifier
5.3. Muscle Synergy Based Classification Is a Potential Method for Knee Pathology Diagnosis
5.4. Limitation and Future Work
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martinez-Villasenor, L.; Ponce, H. Design and Analysis for Fall Detection System Simplification. J. Vis. Exp. 2020, 158. [Google Scholar] [CrossRef]
- Xi, X.; Tang, M.; Miran, S.M.; Luo, Z. Evaluation of Feature Extraction and Recognition for Activity Monitoring and Fall Detection Based on Wearable sEMG Sensors. Sensors 2017, 17, 1229. [Google Scholar] [CrossRef]
- Zhang, Z.; Sup, F. Activity recognition of the torso based on surface electromyography for exoskeleton control. Biomed. Signal Process. Control 2014, 10, 281–288. [Google Scholar] [CrossRef]
- Duan, F.; Dai, L.L.; Chang, W.N.; Chen, Z.Q.; Zhu, C.; Li, W. sEMG-Based Identification of Hand Motion Commands Using Wavelet Neural Network Combined With Discrete Wavelet Transform. IEEE Trans. Ind. Electron. 2016, 63, 1923–1934. [Google Scholar] [CrossRef]
- Kumari, P.; Mathew, L.; Syal, P. Increasing trend of wearables and multimodal interface for human activity monitoring: A review. Biosens. Bioelectron. 2017, 90, 298–307. [Google Scholar] [CrossRef]
- Roy, S.H.; Cheng, M.S.; Chang, S.S.; Moore, J.; De Luca, G.; Nawab, S.H.; De Luca, C.J. A Combined sEMG and Accelerometer System for Monitoring Functional Activity in Stroke. IEEE Trans. Neural Syst. Rehabil. Eng. 2009, 17, 585–594. [Google Scholar] [CrossRef]
- Rafiee, J.; Rafiee, M.A.; Yavari, F.; Schoen, M.P. Feature extraction of forearm EMG signals for prosthetics. Expert Syst. Appl. 2011, 38, 4058–4067. [Google Scholar] [CrossRef]
- Khushaba, R.N.; Kodagoda, S.; Takruri, M.; Dissanayake, G. Toward improved control of prosthetic fingers using surface electromyogram (EMG) signals. Expert Syst. Appl. 2012, 39, 10731–10738. [Google Scholar] [CrossRef]
- Joshi, C.D.; Lahiri, U.; Thakor, N.V. Classification of Gait Phases from Lower Limb EMG: Application to Exoskeleton Orthosis. In Proceedings of the 2013 IEEE Point-of-Care Healthcare Technologies (PHT), Bangalore, India, 16–18 January 2013; pp. 228–231. [Google Scholar]
- Ericson, M.O.; Nisell, R.; Ekholm, J. Quantified electromyography of lower-limb muscles during level walking. Scand. J. Rehabil. Med. 1986, 18, 159–163. [Google Scholar]
- Murley, G.S.; Menz, H.B.; Landorf, K.B.; Bird, A.R. Reliability of lower limb electromyography during overground walking: A comparison of maximal- and sub-maximal normalisation techniques. J. Biomech. 2010, 43, 749–756. [Google Scholar] [CrossRef]
- Rainoldi, A.; Melchiorri, G.; Caruso, I. A method for positioning electrodes during surface EMG recordings in lower limb muscles. J. Neurosci. Methods 2004, 134, 37–43. [Google Scholar] [CrossRef]
- Bernstein, N.A. The Coordination and Regulation Of Movements; Pergamon: New York, NY, USA, 1967. [Google Scholar]
- Maassen; Ben; Lieshout, V. Motor Control: Translating Research into Clinical Practice; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- D’Avella, A.; Saltiel, P.; Bizzi, E. Combinations of muscle synergies in the construction of a natural motor behavior. Nat. Neurosci. 2003, 6, 300–308. [Google Scholar] [CrossRef]
- Ting, L.H.; McKay, J.L. Neuromechanics of muscle synergies for posture and movement. Curr. Opin. Neurobiol. 2007, 17, 622–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berniker, M.; Jarc, A.; Bizzi, E.; Tresch, M.C. Simplified and effective motor control based on muscle synergies to exploit musculoskeletal dynamics. Proc. Natl. Acad. Sci. USA 2009, 106, 7601–7606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neptune, R.R.; Clark, D.J.; Kautz, S.A. Modular control of human walking: A simulation study. J. Biomech. 2009, 42, 1282–1287. [Google Scholar] [CrossRef] [Green Version]
- Naik, G.R.; Selvan, S.E.; Arjunan, S.P.; Acharyya, A.; Kumar, D.K.; Ramanujam, A.; Nguyen, H.T. An ICA-EBM-Based sEMG Classifier for Recognizing Lower Limb Movements in Individuals With and Without Knee Pathology. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Algorithms for non-negative matrix factorization. In Advances in Neural Information Processing Systems 13 (NIPS 2000), Denver, USA; Leen, T.K., Dietterich, T.G., Tresp, V., Eds.; The MIT Press: Cambridge, MA, USA, 2001; Volume 13, pp. 556–562. [Google Scholar]
- Naik, G.R.; Nguyen, H.T. Nonnegative Matrix Factorization for the Identification of EMG Finger Movements: Evaluation Using Matrix Analysis. IEEE J. Biomed. Health Inform. 2015, 19, 478–485. [Google Scholar] [CrossRef]
- Chen, X.; Niu, X.C.; Wu, D.; Yu, Y.; Zhang, X. Investigation of the Intra- and Inter-Limb Muscle Coordination of Hands-and-Knees Crawling in Human Adults by Means of Muscle Synergy Analysis. Entropy 2017, 19, 229. [Google Scholar] [CrossRef]
- Hagio, S.; Fukuda, M.; Kouzaki, M. Identification of muscle synergies associated with gait transition in humans. Front. Hum. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [Green Version]
- Saito, A.; Tomita, A.; Ando, R.; Watanabe, K.; Akima, H. Muscle synergies are consistent across level and uphill treadmill running. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Ozaki, Y.; Aoki, R.; Kimura, T.; Takashima, Y.; Yamada, T. Characterizing Muscular Activities Using Non-negative Matrix Factorization from EMG Channels for Driver Swings in Golf. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 892–895. [Google Scholar]
- Tran, B.N.; Yano, S.; Kondo, T. Muscle Synergy Analysis in Dart Throwing. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 2534–2537. [Google Scholar]
- Smale, K.B.; Shourijeh, M.S.; Benoit, D.L. Use of muscle synergies and wavelet transforms to identify fatigue during squatting. J. Electromyogr. Kinesiol. 2016, 28, 158–166. [Google Scholar] [CrossRef]
- Rodriguez, K.L.; Roemmich, R.T.; Cam, B.; Fregly, B.J.; Hass, C.J. Persons with Parkinson’s disease exhibit decreased neuromuscular complexity during gait. Clin. Neurophysiol. 2013, 124, 1390–1397. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Chen, X.; Cao, S.; Wu, D.; Zhao, G.; Zhang, X. Assessment of Upper Limb Motor Dysfunction for Children with Cerebral Palsy Based on Muscle Synergy Analysis. Front. Hum. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [Green Version]
- Hayes, H.B.; Chvatal, S.A.; French, M.A.; Ting, L.N.H.; Trumbower, R.D. Neuromuscular constraints on muscle coordination during overground walking in persons with chronic incomplete spinal cord injury. Clin. Neurophysiol. 2014, 125, 2024–2035. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Wang, B.H.; Jiang, N.; Farina, D. Robust extraction of basis functions for simultaneous and proportional myoelectric control via sparse non-negative matrix factorization. J. Neural Eng. 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Mathieu, P.A. Static hand posture classification based on the biceps brachii muscle synergy features. J. Electromyogr. Kinesiol. 2018, 43, 201–208. [Google Scholar] [CrossRef]
- Israely, S.; Leisman, G.; Machluf, C.C.; Carmeli, E. Muscle Synergies Control during Hand-Reaching Tasks in Multiple Directions Post-stroke. Front. Comput. Neurosci. 2018, 12. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.Y.; Wu, X.Y.; Chen, L.; Zhao, Y.; Zhang, L.; Li, G.L.; Hou, W.S. Synergistic Myoelectrical Activities of Forearm Muscles Improving Robust Recognition of Multi-Fingered Gestures. Sensors 2019, 19, 610. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Xian, Z.E.; Tang, F.; Li, L.L.; Zhang, L.; Zhang, Z.G. Low-density surface electromyographic patterns under electrode shift: Characterization and NMF-based classification. Biomed. Signal Process. Control 2020, 59. [Google Scholar] [CrossRef]
- Qiao, H.L. New SVD based initialization strategy for non-negative matrix factorization. Pattern Recognit. Lett. 2015, 63, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Saito, A.; Tomita, A.; Ando, R.; Watanabe, K.; Akima, H. Similarity of muscle synergies extracted from the lower limb including the deep muscles between level and uphill treadmill walking. Gait Posture 2017, 59, 134. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, O.F.A.; Sotelo, J.L.R.; Gonzales, M.H.; Hernandez, G.A.M. UCI Machine Learning Repository; University of California, School of Information and Computer Science: Irvine, CA, USA, 2014. [Google Scholar]
- Herrera-González, M.; Rodríguez-Sotelo, J.; Avilés-Sánchez, Ó. Knee Functional State Classification Using Surface Electromyographic and Goniometric Signals by Means of Artificial Neural Networks. J. Chem. Technol. Biotechnol. 2015, 87, 1385–1395. [Google Scholar]
- Israely, S.; Leisman, G.; Machluf, C.; Shnitzer, T.; Carmeli, E. Direction Modulation of Muscle Synergies in a Hand-Reaching Task. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 2427–2440. [Google Scholar] [CrossRef]
- He, T.; Hu, J.; Xia, P.; Gu, C. Feature Selection of Emg Signal Based on ReliefF Algorithm and Genetic Algorithm. J. Shanghai Jiaotong Univ. 2016, 50, 204–208. [Google Scholar] [CrossRef]
- Benazzouz, A.; Guilal, R.; Amirouche, F.; Hadj Slimane, Z.E. EMG Feature Selection for Diagnosis of Neuromuscular Disorders. In Proceedings of the 2019 International Conference on Networking and Advanced Systems (ICNAS), Annaba, Algeria, 26–27 June 2019. [Google Scholar]
- Altun, K.; Barshan, B. Human Activity Recognition Using Inertial/Magnetic Sensor Units. In Human Behavior Understanding; Salah, A.A., Gevers, T., Sebe, N., Vinciarelli, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6219, pp. 38–51. [Google Scholar]
- Jia, R.; Liu, B. Human daily activity recognition by fusing accelerometer and multi-lead ECG data. In Proceedings of the 2013 IEEE International Conference on Signal Processing, Communication and Computing (ICSPCC 2013), KunMing, China, 5–8 August 2013. [Google Scholar]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008. [Google Scholar]
- Chen, J.C.; Sun, Y.N.; Sun, S.M. Improving Human Activity Recognition Performance by Data Fusion and Feature Engineering. Sensors 2021, 21, 692. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Scorneti, E. Tuning parameters in random forests. ESAIM Proc. Surv. 2017, 60, 144–162. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, H.; Ogawa, T.; Kawashima, N.; Shinya, M.; Nakazawa, K. Distinct sets of locomotor modules control the speed and modes of human locomotion. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Auriol, P.A.; Besier, T.F.; Byblow, W.D.; Mcmorland, A. Fatigue Influences the Recruitment, but Not Structure, of Muscle Synergies. Front. Hum. Neurosci. 2018, 12, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunardini, F.; Casellato, C.; Bertucco, M.; Sanger, T.D.; Pedrocchi, A. Children with and without Dystonia Share Common Muscle Synergies While Performing Writing Tasks. Ann. Biomed. Eng. 2017, 45, 1–14. [Google Scholar] [CrossRef]
- Janidarmian, M.; Radecka, K.; Zilic, Z. Automated Diagnosis of Knee Pathology Using Sensory Data. In Proceedings of the 2014 4th International Conference on Wireless Mobile Communication and Healthcare—Transforming Healthcare Through Innovations in Mobile and Wireless Technologies (MOBIHEALTH), Athens, Greece, 3–5 November 2014; pp. 95–98. [Google Scholar]
- Ertugrul, O.F.; Kaya, Y.; Tekin, R. A novel approach for SEMG signal classification with adaptive local binary patterns. Med. Biol. Eng. Comput. 2016, 54, 1137–1146. [Google Scholar] [CrossRef] [PubMed]












| Motion | Number of Synergy | Control Group | Study Group |
|---|---|---|---|
| Leg flexion at standing position (STD) | 1 | 0.7388 | 0.8015 |
| 2 | 0.8570 | 0.9058 | |
| 3 | 0.9500 | 0.9725 | |
| Leg extension at sitting position (ST) | 1 | 0.8088 | 0.6633 |
| 2 | 0.8989 | 0.8730 | |
| 3 | 0.9580 | 0.9338 | |
| Gait exercise (Gait) | 1 | 0.6486 | 0.5797 |
| 2 | 0.8085 | 0.7586 | |
| 3 | 0.8973 | 0.8639 |
| Feature Sets | Subject Group | k = 2 | k = 3 |
|---|---|---|---|
| Coarse Feature Sets | Control Group | 0.920 | 0.902 |
| Study Group | 0.938 | 0.910 | |
| Combined Group | 0.913 | 0.897 | |
| Fine Feature Sets | Control Group | 0.968 | 0.955 |
| Study Group | 0.973 | 0.958 | |
| Combined Group | 0.962 | 0.952 | |
| Selected Feature Sets | Control Group | 0.964 | 0.944 |
| Study Group | 0.973 | 0.949 | |
| Combined Group | 0.960 | 0.943 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Sun, Y.; Sun, S. Muscle Synergy of Lower Limb Motion in Subjects with and without Knee Pathology. Diagnostics 2021, 11, 1318. https://doi.org/10.3390/diagnostics11081318
Chen J, Sun Y, Sun S. Muscle Synergy of Lower Limb Motion in Subjects with and without Knee Pathology. Diagnostics. 2021; 11(8):1318. https://doi.org/10.3390/diagnostics11081318
Chicago/Turabian StyleChen, Jingcheng, Yining Sun, and Shaoming Sun. 2021. "Muscle Synergy of Lower Limb Motion in Subjects with and without Knee Pathology" Diagnostics 11, no. 8: 1318. https://doi.org/10.3390/diagnostics11081318





