Evaluation of Hepatitis C Virus Core Antigen Assay in a Resource-Limited Setting in Pakistan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Decentralized Lab Set-Up
2.3. Study Procedures
2.4. Study Assessment
2.5. Statistical Analysis
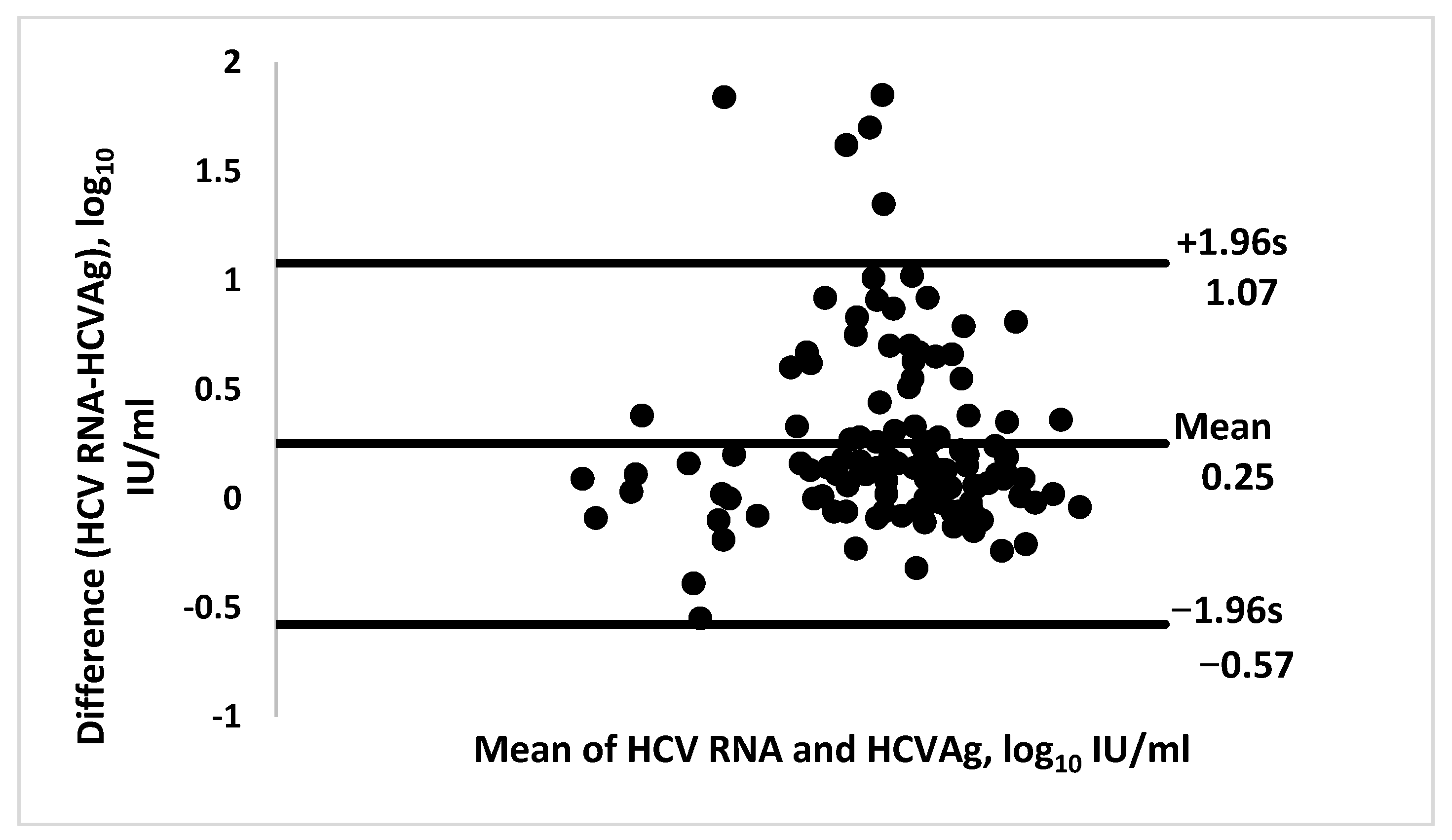
3. Results
4. Operational Issues
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stanaway, J.D.; Flaxman, A.D.; Naghavi, M.; Fitzmaurice, C.; Vos, T.; Abubakar, I.; Abu-Raddad, L.J.; Assadi, R.; Bhala, N.; Cowie, B.; et al. The global burden of viral hepatitis from 1990 to 2013: Findings from the Global Burden of Disease Study 2013. Lancet 2016, 388, 1081–1088. [Google Scholar] [CrossRef] [Green Version]
- Blach, S.; Zeuzem, S.; Manns, M.; Altraif, I.; Duberg, A.-S.; Muljono, D.H.; Waked, I.; Alavian, S.M.; Lee, M.-H.; Negro, F.; et al. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: A modelling study. Lancet Gastroenterol. Hepatol. 2017, 2, 161–176. [Google Scholar] [CrossRef] [Green Version]
- Grebely, J.; Applegate, T.; Cunningham, P.; Feld, J.J. Hepatitis C point-of-care diagnostics: In search of a single visit diagnosis. Expert Rev. Mol. Diagn. 2017, 17, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Applegate, T.L.; Fajardo, E.; Sacks, J.A. Hepatitis C Virus Diagnosis and the Holy Grail. Infect. Dis. Clin. N. Am. 2018, 32, 425–445. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, H.; Bile, K.; Jooma, R.; Alam, S.; Afrid, H. Prevalence of hepatitis B and C viral infections in Pakistan: Findings of a national survey appealing for effective prevention and control measures. East. Mediterr. Health J. 2010, 16, 15–23. [Google Scholar] [CrossRef]
- Population Census|Pakistan Bureau of Statistics. Available online: http://www.pbs.gov.pk/content/population-census (accessed on 13 January 2021).
- Janjua, N.Z.; Hamza, H.B.; Islam, M.; Tirmizi, S.F.A.; Siddiqui, A.; Jafri, W.; Hamid, S. Health care risk factors among women and personal behaviours among men explain the high prevalence of hepatitis C virus infection in Karachi, Pakistan. J. Viral. Hepat. 2010, 17, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Catlett, B.; Lamoury, F.M.J.; Bajis, S.; Hajarizadeh, B.; Martinez, D.; Mowat, Y.; Cunningham, P.H.; Jacka, B.P.; A Cloherty, G.; Marks, P.; et al. Evaluation of a hepatitis C virus core antigen assay from venepuncture and dried blood spot collected samples: A cohort study. J. Viral Hepat. 2019, 26, 1423–1430. [Google Scholar] [CrossRef]
- Cohn, J.; Roberts, T.; Amorosa, V.; Lemoine, M.; Hill, A. Simplified diagnostic monitoring for hepatitis C, in the new era of direct-acting antiviral treatment. Curr. Opin. HIV AIDS 2015, 10, 369–373. [Google Scholar] [CrossRef]
- Lamoury, F.M.; Hajarizadeh, B.; Soker, A.; Martinez, D.; Quek, C.; Cunningham, P.; Catlett, B.; Cloherty, G.; Marks, P.; Amin, J.; et al. Evaluation of a Hepatitis C Virus Core Antigen Assay in Plasma and Dried Blood Spot Samples. J. Mol. Diagn. 2018, 20, 621–627. [Google Scholar] [CrossRef]
- Lemoine, M.; Lacombe, K. HCV core antigen: Toward elimination of nucleic acid testing? Lancet Gastroenterol. Hepatol. 2018, 3, 817–818. [Google Scholar] [CrossRef]
- Freiman, J.M.; Tran, T.M.; Schumacher, S.G.; White, L.F.; Ongarello, S.; Cohn, J.; Easterbrook, P.J.; Linas, B.P.; Denkinger, C.M. Hepatitis C core antigen testing for diagnosis of hepatitis C virus infection: A systematic review and meta-analysis. Ann. Intern. Med. 2016, 165, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Galli, C.; Julicher, P.; Plebani, M. HCV core antigen comes of age: A new opportunity for the diagnosis of hepatitis C virus infection. Clin. Chem. Lab. Med. 2018, 56, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Van Tilborg, M.; Al Marzooqi, S.H.; Wong, W.W.; Maan, R.; Vermehren, J.; Maasoumy, B.; Mazzulli, T.; Bolotin, S.; Garber, G.; Guerra, F. HCV core antigen as an alternative to HCV RNA testing in the era of direct-acting antivirals: Retrospective screening and diagnostic cohort studies. Lancet Gastroenterol. Hepatol. 2018, 3, 856–864. [Google Scholar] [CrossRef]
- Mohamed, Z.; Mbwambo, J.; Shimakawa, Y.; Poiteau, L.; Chevaliez, S.; Pawlotsky, J.-M.; Rwegasha, J.; Bhagani, S.; Taylor-Robinson, S.D.; Makani, J.; et al. Clinical utility of HCV core antigen detection and quantification using serum samples and dried blood spots in people who inject drugs in Dar-es-Salaam, Tanzania. J. Int. AIDS Soc. 2017, 20, 1–7. [Google Scholar] [CrossRef]
- Çetiner, S.; Duran, A.Ç.; Kibar, F.; Yaman, A. Performance comparison of new generation HCV core antigen test versus HCV RNA test in management of hepatitis C virus infection. Transfus. Apher. Sci. 2017, 56, 362–366. [Google Scholar] [CrossRef]
- Lamoury, F.M.; Soker, A.; Martinez, D.; Hajarizadeh, B.; Cunningham, E.B.; Cunningham, P.; Bruggmann, P.; Foster, G.R.; Dalgard, O.; Backmund, M.; et al. Hepatitis C virus core antigen: A simplified treatment monitoring tool, including for post-treatment relapse. J. Clin. Virol. 2017, 92, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Wakeel, M.; Chen, B.; Jahangir, S. Overview of Energy Portfolio in Pakistan. Energy Procedia 2016, 88, 71–75. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Pérez-García, F.; Ampuero, D.; Reigadas, E.; Bouza, E. New direct-acting antivirals for patients with chronic HCV infection: Can we monitor treatment using an HCV core antigen assay? Diagn. Microbiol. Infect. Dis. 2017, 87, 243–246. [Google Scholar] [CrossRef]
- Lamoury, F.M.J.; Bajis, S.; Hajarizadeh, B.; Marshall, A.; Martinello, M.; Ivanova, E.; Catlett, B.; Mowat, Y.; Marks, P.; Amin, J.; et al. Evaluation of the Xpert HCV Viral Load Finger-Stick Point-of-Care Assay. J. Infect. Dis. 2018, 217, 1889–1896. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.; Roberts, T.; Cohn, J.; Greenman, J.; Camp, J.; Ishizaki, A.; Messac, L.; Tuaillon, E.; Van de Perre, P.; Pichler, C.; et al. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples—A systematic review and meta-analysis. BMC Infect. Dis. 2017, 17, 71–85. [Google Scholar] [CrossRef] [Green Version]
- EASL Recommendations on Treatment of Hepatitis C 2016. J. Hepatol. 2017, 66, 153–194. [CrossRef] [Green Version]
- Duchesne, L.; Hejblum, G.; Njouom, R.; Kane, C.T.; Toni, T.D.; Moh, R.; Sylla, B.; Rouveau, N.; Attia, A.; Lacombe, K. Model-based cost-effectiveness estimates of testing strategies for diagnosing hepatitis C virus infection in Central and Western Africa. PLoS ONE 2020, 15, e0238035. [Google Scholar] [CrossRef]
- Chevaliez, S.; Feld, J.; Cheng, K.; Wedemeyer, H.; Sarrazin, C.; Maasoumy, B.; Herman, C.; Hackett, J.; Cohen, D.; Dawson, G.; et al. Clinical utility of HCV core antigen detection and quantification in the diagnosis and management of patients with chronic hepatitis C receiving an all-oral, interferon-free regimen. Antivir Ther. 2016, 23, 211–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockstroh, J.K.; Feld, J.J.; Chevaliez, S.; Cheng, K.; Wedemeyer, H.; Sarrazin, C.; Maasoumy, B.; Herman, C.; Hackett, J.; Cohen, D.E.; et al. HCV core antigen as an alternate test to HCV RNA for assessment of virologic responses to all-oral, interferon-free treatment in HCV genotype 1 infected patients. J. Virol. Methods 2017, 245, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Boeras, D.I.; Marinucci, F.; Easterbrook, P. The future of viral hepatitis testing: Innovations in testing technologies and approaches. BMC Infect. Dis. 2017, 17, 187–196. [Google Scholar] [CrossRef] [PubMed]

| HCVcAg Assay (fmol/L) | HCV RNA (IU/mL) | |
|---|---|---|
| 1 | 3.08 (Reactive) | Not Detected Ω |
| 2 | 4.61 (Reactive) | Not Detected Ω |
| 3 | 3.00 (Reactive) | Not Detected Ω |
| 4 | 10.44 (Reactive) | Not Detected Ω |
| 5 | 0.00 (Not Reactive) | 41 (Detected) |
| 6 | 1.68 (Not Reactive) | 669 (Detected) |
| 7 | 2.56 (Not Reactive) | 90100 (Detected) |
| 8 | 1.02 (Not Reactive) | 638 (Detected) |
| 9 | 2.53 (Not Reactive) | 2990 (Detected) |
| 10 | 0.00 (Not Reactive) | 411 (Detected) |
| Reactive | Not Reactive | Total | Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|---|---|---|
| HCV RNA Plasma (≥12 IU/mL) | |||||
| Detected | 122 | 6 | 128 | 96.8% (92–99%) | 91.8% (83–97%) |
| Undetected | 4 | 68 | 72 | ||
| Total | 126 | 74 | 200 | ||
| HCV RNA Plasma (≥1000 IU/mL) | |||||
| Detected | 121 | 2 | 123 | 96.0% (91–99%) | 97.2% (90–100%) |
| Undetected | 5 | 72 | 77 | ||
| HCV RNA Plasma (≥3000 IU/mL) | |||||
| Detected | 119 | 1 | 120 | 94.4% (88–98%) | 98.6% (92–100%) |
| Undetected | 7 | 73 | 80 | ||
| Reactive | Not Reactive | Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|---|---|
| HCV RNA Plasma (≥12 IU/mL) | ||||
| Detected | 118 | 10 | 99.1% (95–100%) | 87.6% (78.4–94%) |
| Undetected | 1 | 71 | ||
| HCV RNA Plasma (≥1000 IU/mL) | ||||
| Detected | 118 | 5 | 99.1% (95–100%) | 93.8% (86–98%) |
| Undetected | 1 | 76 | ||
| HCV RNA Plasma (≥3000 IU/mL) | ||||
| Detected | 118 | 2 | 99.1% (95–100%) | 97.5% (91–100%) |
| Undetected | 1 | 79 | ||
| Reactive | Not Reactive | Observed Agreement | ĸ Statistic | |
|---|---|---|---|---|
| HCV core antigen plasma (≥3 fmol/L) | ||||
| HCV RNA Plasma (≥12 IU/mL) | ||||
| Detected | 122 | 6 | 190/200 | 0.90 |
| Undetected | 4 | 68 | ||
| HCV RNA Plasma (≥1000 IU/mL) | ||||
| Detected | 121 | 2 | 193/200 | 0.93 |
| Undetected | 5 | 72 | ||
| HCV RNA Plasma (≥3000 IU/mL) | ||||
| Detected | 119 | 1 | 192/200 | 0.92 |
| Undetected | 7 | 73 | ||
| HCV core antigen plasma (≥10 fmol/L) | ||||
| HCV RNA Plasma (≥12 IU/mL) | ||||
| Detected | 118 | 10 | 189/200 | 0.88 |
| Undetected | 1 | 71 | ||
| HCV RNA Plasma (≥1000 IU/mL) | ||||
| Detected | 118 | 5 | 194/200 | 0.93 |
| Undetected | 1 | 76 | ||
| HCV RNA Plasma (≥3000 IU/mL) | ||||
| Detected | 118 | 2 | 197/200 | 0.96 |
| Undetected | 1 | 79 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abid, A.; Uddin, M.; Muhammad, T.; Awan, S.; Applegate, T.; Dore, G.J.; Cloherty, G.; Hamid, S. Evaluation of Hepatitis C Virus Core Antigen Assay in a Resource-Limited Setting in Pakistan. Diagnostics 2021, 11, 1354. https://doi.org/10.3390/diagnostics11081354
Abid A, Uddin M, Muhammad T, Awan S, Applegate T, Dore GJ, Cloherty G, Hamid S. Evaluation of Hepatitis C Virus Core Antigen Assay in a Resource-Limited Setting in Pakistan. Diagnostics. 2021; 11(8):1354. https://doi.org/10.3390/diagnostics11081354
Chicago/Turabian StyleAbid, Adeel, Murad Uddin, Taj Muhammad, Safia Awan, Tanya Applegate, Gregory J. Dore, Gavin Cloherty, and Saeed Hamid. 2021. "Evaluation of Hepatitis C Virus Core Antigen Assay in a Resource-Limited Setting in Pakistan" Diagnostics 11, no. 8: 1354. https://doi.org/10.3390/diagnostics11081354
APA StyleAbid, A., Uddin, M., Muhammad, T., Awan, S., Applegate, T., Dore, G. J., Cloherty, G., & Hamid, S. (2021). Evaluation of Hepatitis C Virus Core Antigen Assay in a Resource-Limited Setting in Pakistan. Diagnostics, 11(8), 1354. https://doi.org/10.3390/diagnostics11081354







