Promoter Hypermethylation of Tumor Suppressor Genes Located on Short Arm of the Chromosome 3 as Potential Biomarker for the Diagnosis of Nasopharyngeal Carcinoma
Abstract
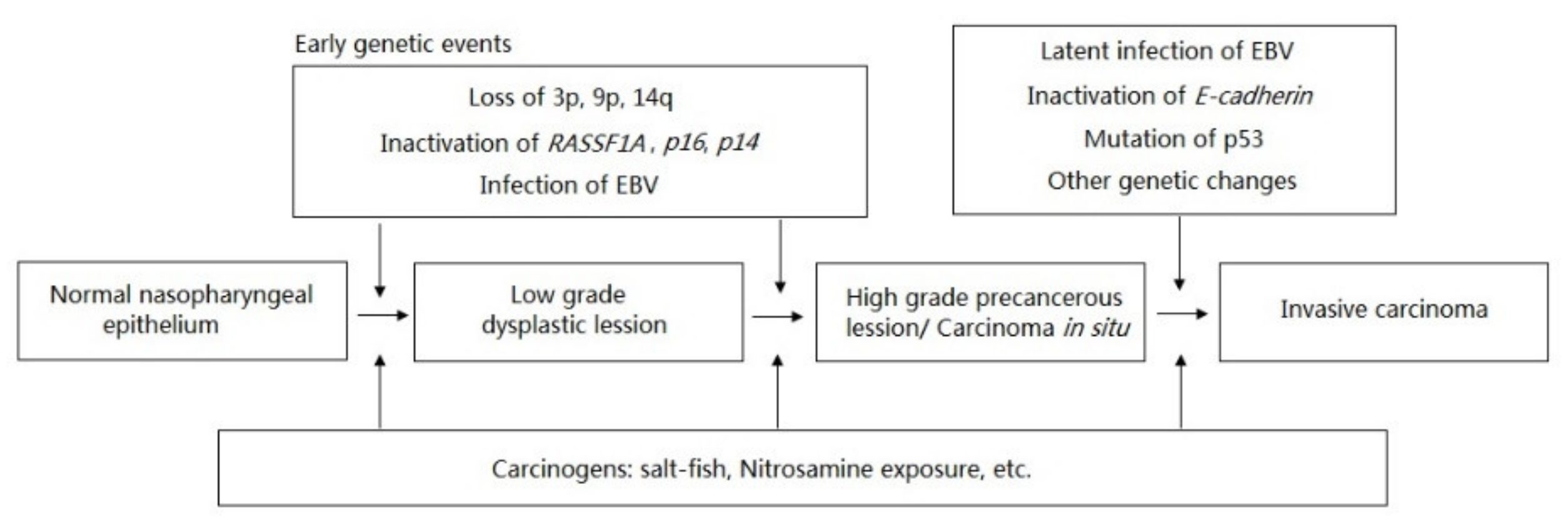
1. Introduction
2. The Inactivation of Tumor Suppressor Genes on Chromosome 3
3. RASSF1A and Panel of Genes as Methylation Biomarkers for NPC
4. Other Genes Located at 3p Have Also Been Studied though Not as much as RASSF1A
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Tham, I.W.K.; Lu, J.J. Treatment of early stage nasopharyngeal carcinoma: Conventional versus new radiation therapy technologies. J. Radiat. Oncol. 2012, 1, 99–106. [Google Scholar] [CrossRef][Green Version]
- Tsao, S.W.; Yip, Y.L.; Tsang, C.M.; Pang, P.S.; Lau, V.M.Y.; Zhang, G.; Lo, K.W. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014, 50, 330–338. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2009, 31, 27–36. [Google Scholar] [CrossRef]
- Hutajulu, S.H.; Indrasari, S.R.; Indrawati, L.P.L.; Harijadi, A.; Duin, S.; Haryana, S.M.; Steenbergen, R.D.M.; Greijer, A.E.; Middeldorp, J.M. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol. Cancer 2011, 10, 48. [Google Scholar] [CrossRef]
- Hong, L.L.; Lo, P.H.Y.; Xie, D.; Apte, S.S.; Cheung, A.K.L.; Cheng, Y.; Law, E.W.L.; Chua, D.; Zeng, Y.X.; Sai, W.T.; et al. Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int. J. Cancer 2008, 123, 401–408. [Google Scholar] [CrossRef]
- Ayadi, W.; Karray-Hakim, H.; Khabir, A.; Feki, L.; Charfi, S.; Boudawara, T.; Ghorbel, A.; Daoud, J.; Frikha, M.; Busson, P.; et al. Aberrant methylation of p16, DLEC1, BLU and E-cadherin gene promoters in nasopharyngeal carcinoma biopsies from Tunisian patients. Anticancer Res. 2008, 28, 2161–2167. [Google Scholar] [PubMed]
- Waddington, C.H. The Epigenotype. Int. J. Epidemiol. 2012, 41, 10–13. [Google Scholar] [CrossRef]
- Han, B.; Yang, X.; Zhang, P.; Zhang, Y.; Tu, Y.; He, Z.; Li, Y.; Yuan, J.; Dong, Y.; Hosseini, D.K.; et al. DNA methylation biomarkers for nasopharyngeal carcinoma. PLoS ONE 2020, 15, 1–16. [Google Scholar] [CrossRef]
- Thuy, L.H.A.; Thuan, L.D.; Phuong, T.K. DNA Hypermethylation in Breast Cancer. In Breast Cancer-From Biology to Medicine; InTech: London, UK, 2017. [Google Scholar]
- Hui, A.B.-Y.; Lo, K.-W.; Leung, S.-F.; Teo, P.; Fung, M.K.F.; To, K.F.; Wong, N.; Choi, P.H.K.; Lee, J.C.K.; Huang, D.P. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int. J. Cancer 1999, 82, 498–503. [Google Scholar] [CrossRef]
- Li, X.; Wang, E.; Zhao, Y.; Ren, J.-Q.; Jin, P.; Yao, K.-T.; Marincola, F.M. Chromosomal imbalances in nasopharyngeal carcinoma: A meta-analysis of comparative genomic hybridization results. J. Transl. Med. 2006, 4, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chow, L.S.N.; Lo, K.W.; Kwong, J.; To, K.F.; Tsang, K.S.; Lam, C.W.; Dammann, R.; Huang, D.P. RASSF1A is a target tumor suppressor from 3p21.3 in nasopharyngeal carcinoma. Int. J. Cancer 2004, 109, 839–847. [Google Scholar] [CrossRef]
- Chen, J.; Fu, L.; Zhang, L.Y.; Kwong, D.L.; Yan, L.; Guan, X.Y. Tumor suppressor genes on frequently deleted chromosome 3p in nasopharyngeal carcinoma. Chin. J. Cancer 2012, 31, 215–222. [Google Scholar] [CrossRef]
- Nawaz, I.; Hu, L.F.; Du, Z.M.; Moumad, K.; Ignatyev, I.; Pavlova, T.V.; Kashuba, V.; Almgren, M.; Zabarovsky, E.R.; Ernberg, I. Integrin α9 gene promoter is hypermethylated and downregulated in nasopharyngeal carcinoma. Oncotarget 2015, 6, 31493–31507. [Google Scholar] [CrossRef] [PubMed]
- Wing, L.Y.; Hong, L.L.; Zabarovsky, E.R.; Lerman, M.I.; Sham, J.S.T.; Chua, D.T.T.; Sai, W.T.; Stanbridge, E.J.; Lung, M.L. Functional studies of the chromosome 3p21.3 candidate tumor suppressor gene BLU/ZMYND10 in nasopharyngeal carcinoma. Int. J. Cancer 2006, 119, 2821–2826. [Google Scholar] [CrossRef]
- Qiu, G.H.; Tan, L.K.S.; Loh, K.S.; Lim, C.Y.; Srivastava, G.; Tsai, S.T.; Tsao, S.W.; Tao, Q. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21.3, is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene 2004, 23, 4793–4806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leung Cheung, A.K.; Lung, H.L.; Hung, S.C.; Law, E.W.L.; Cheng, Y.; Yau, W.L.; Bangarusamy, D.K.; Miller, L.D.; Liu, E.T.B.; Shao, J.Y.; et al. Functional analysis of a cell cycle-associated, tumor-suppressive gene, protein tyrosine phosphatase receptor type G, in nasopharyngeal carcinoma. Cancer Res. 2008, 68, 8137–8145. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Q.; Chen, H.K.; Zhang, X.S.; Pan, Z.G.; Li, A.; Feng, Q.S.; Long, Q.X.; Wang, X.Z.; Zeng, Y.X. Alterations of BLU, a candidate tumor suppressor gene on chromosome 3p21.3, in human nasopharyngeal carcinoma. Int. J. Cancer 2003, 106, 60–65. [Google Scholar] [CrossRef]
- Lao, T.D.; Huyen Le, T.A. Characteristic of ZMYND10 Gene’s Promoter Hypermethylation in Nasopharyngeal Carcinoma Biopsies from Vietnamese Patients. Asian J. Pharm. Res. Health Care 2019, 10, 60–65. [Google Scholar] [CrossRef]
- Yi, H.-M.; Ren, C.-P.; Peng, D.; Zhou, L.; Li, H.; Yao, K.-T. [Expression, loss of heterozygosity, and methylation of GNAT1 gene in nasopharyngeal carcinoma]. Ai Zheng 2007, 26, 9–14. [Google Scholar]
- Zhou, W.; Feng, X.; Li, H.; Wang, L.; Zhu, B.; Liu, W.; Zhao, M.; Yao, K.; Ren, C. Inactivation of LARS2, located at the commonly deleted region 3p21.3, by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Acta Biochim. Biophys. Sin. 2009, 41, 54–62. [Google Scholar] [CrossRef]
- Yi, H.-M.; Li, H.; Peng, D.; Zhang, H.-J.; Wang, L.; Zhao, M.; Yao, K.-T.; Ren, C.-P. Genetic and epigenetic alterations of LTF at 3p21.3 in nasopharyngeal carcinoma. Oncol. Res. 2006, 16, 261–272. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, X.; Liu, W.; Jiang, X.; Shan, W.; Huang, C.; Yi, H.; Zhu, B.; Zhou, W.; Wang, L.; et al. Underlying mechanisms for LTF inactivation and its functional analysis in nasopharyngeal carcinoma cell lines. J. Cell. Biochem. 2011, 112, 1832–1843. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, W.; Tang, H.; Ye, Q.; Liao, Q.; Zhou, Y.; Wu, M.; Xiong, W.; Zheng, Y.; Guo, X.; et al. Lactotransferrin acts as a tumor suppressor in nasopharyngeal carcinoma by repressing AKT through multiple mechanisms. Oncogene 2013, 32, 4273–4283. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Kwong, D.L.W.; Sham, J.S.T.; Wei, W.I.; Kwong, Y.L.; Yuen, A.P.W. Quantitative Plasma Hypermethylated DNA Markers of Undifferentiated Nasopharyngeal Carcinoma. Clin. Cancer Res. 2004, 10, 2401–2406. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Tang, K.C.; Kwong, D.L.W.; Sham, J.S.T.; Wei, W.I.; Kwong, Y.L.; Yuen, A.P.W. Differential gene methylation in undifferentiated nasopharyngeal carcinoma. Int. J. Oncol. 2003, 22, 869–874. [Google Scholar] [CrossRef]
- Challouf, S.; Ziadi, S.; Zaghdoudi, R.; Ksiaa, F.; Ben Gacem, R.; Trimeche, M. Patterns of aberrant DNA hypermethylation in nasopharyngeal carcinoma in Tunisian patients. Clin. Chim. Acta 2012, 413, 795–802. [Google Scholar] [CrossRef]
- Chang, H.W.; Chan, A.; Kwong, D.L.W.; Wei, W.I.; Suam, J.S.T.; Yuen, A.P.W. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int. J. Cancer 2003, 105, 851–855. [Google Scholar] [CrossRef]
- Kwong, J.; Lo, K.W.; To, K.F.; Teo, P.M.L.; Johnson, P.J.; Poon Huang, D. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin. Cancer Res. 2002, 8, 131–137. [Google Scholar]
- Tong, J.H.M.; Tsang, R.K.Y.; Lo, K.-W.; Woo, J.K.S.; Kwong, J.; Chan, M.W.Y.; Chang, A.R.; van Hasselt, C.A.; Huang, D.P.; To, K.-F. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin. Cancer Res. 2002, 8, 2612–2619. [Google Scholar] [PubMed]
- Zhang, Z.; Sun, D.; Hutajulu, S.H.; Nawaz, I.; Nguyen Van, D.; Huang, G.; Haryana, S.M.; Middeldorp, J.M.; Ernberg, I.; Hu, L.F. Development of a Non-Invasive Method, Multiplex Methylation Specific PCR (MMSP), for Early Diagnosis of Nasopharyngeal Carcinoma. PLoS ONE 2012, 7, 1–6. [Google Scholar] [CrossRef][Green Version]
- Wang, T.; Liu, H.; Chen, Y.; Liu, W.; Yu, J.; Wu, G. Methylation associated inactivation of RASSF1A and its synergistic effect with activated K-Ras in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2009, 28. [Google Scholar] [CrossRef]
- Lo, K.W.; Kwong, J.; Hui, A.B.; Chan, S.Y.; To, K.F.; Chan, A.S.; Chow, L.S.; Teo, P.M.; Johnson, P.J.; Huang, D.P. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001, 61, 3877–3881. [Google Scholar]
- Thieu, H.H.; Lao, T.D.; Le, T.A.H. Characterization of Promoter Hypermethylation of Tumor Suppressor Gene Rassf1a and Its Association With the Risk of Nasopharyngeal Carcinoma. Pharmacophore 2020, 11, 56–62. [Google Scholar]
- Fendri, A.; Masmoudi, A.; Khabir, A.; Sellami-Boudawara, T.; Daoud, J.; Frikha, M.; Ghorbel, A.; Gargouri, A.; Mokdad-Gargouri, R. Inactivation of RASSF1A, RARβ2 and DAP-kinase by promoter methylation correlates with lymph node metastasis in nasopharyngeal carcinoma. Cancer Biol. Ther. 2009, 8, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jiang, W.; Ren, C.; Yin, Z.; Feng, X.; Liu, W.; Taoz, Q.; Yao, K. Frequent hypermethylation of RASSF1A and TSLC1, and high, viral load of Epstein-Barr virus DNA in nasopharyngeal carcinoma and matched tumor-adjacent tissues. Neoplasia 2005, 7, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yip, S.P.; Kwong, D.L.W.; Lin, Z.; Yang, Z.; Wu, V.W.C. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol. 2013, 37, 708–713. [Google Scholar] [CrossRef]
- Loyo, M.; Brait, M.; Kim, M.S.; Ostrow, K.L.; Jie, C.C.; Chuang, A.Y.; Califano, J.A.; Liégeois, N.J.; Begum, S.; Westra, W.H.; et al. A survey of methylated candidate tumor suppressor genes in nasopharyngeal carcinoma. Int. J. Cancer 2011, 128, 1393–1403. [Google Scholar] [CrossRef]
- Kwong, J.; Chow, L.S.-N.; Wong, A.Y.-H.; Hung, W.-K.; Chung, G.T.-Y.; To, K.-F.; Chan, F.L.; Daigo, Y.; Nakamura, Y.; Huang, D.P.; et al. Epigenetic inactivation of the deleted in lung and esophageal cancer 1 gene in nasopharyngeal carcinoma. Genes Chromosom. Cancer 2007, 46, 171–180. [Google Scholar] [CrossRef]
- Kwong, J.; Lo, K.-W.; Chow, L.S.-N.; Chan, F.L.; To, K.-F.; Huang, D.P. Silencing of the retinoid response gene TIG1 by promoter hypermethylation in nasopharyngeal carcinoma. Int. J. Cancer 2005, 113, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.; Li, C.; Yoon, J.-H.; Chin, P.L.; Bates, S.; Pfeifer, G.P. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000, 25, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.W.; To, K.F.; Huang, D.P. Focus on nasopharyngeal carcinoma. Cancer Cell 2004, 5, 423–428. [Google Scholar] [CrossRef]
- Shivakumar, L.; Minna, J.; Sakamaki, T.; Pestell, R.; White, M.A. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol. Cell. Biol. 2002, 22, 4309–4318. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Huang, T.; Ni, C.; Yang, P.; Chen, S. Diagnostic Capacity of RASSF1A Promoter Methylation as a Biomarker in Tissue, Brushing, and Blood Samples of Nasopharyngeal Carcinoma. EBioMedicine 2017, 18, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ooft, M.L.; van Ipenburg, J.; van Loo, R.; de Jong, R.; Moelans, C.; Braunius, W.; de Bree, R.; van Diest, P.; Koljenović, S.; Baatenburg de Jong, R.; et al. Molecular profile of nasopharyngeal carcinoma: Analysing tumour suppressor gene promoter hypermethylation by multiplex ligation-dependent probe amplification. J. Clin. Pathol. 2018, 71, 351–359. [Google Scholar] [CrossRef]
- Zhang, S.-Q.; Pan, S.-M.; Liang, S.-X.; Han, Y.-S.; Chen, H.-B.; Li, J.-C. Research status and prospects of biomarkers for nasopharyngeal carcinoma in the era of high-throughput omics (Review). Int. J. Oncol. 2021, 58, 9. [Google Scholar] [CrossRef]
- Gihbid, A.; Benzeid, R.; Faouzi, A.; Nourlil, J.; Tawfiq, N.; Benchakroun, N.; Guensi, A.; Bendahhou, K.; Benider, A.; El Benna, N.; et al. Circulating cell-free epstein–barr virus DNA levels and clinical features in Moroccan patients with nasopharyngeal carcinoma. Infect. Agent. Cancer 2021, 16, 15. [Google Scholar] [CrossRef]
- Skalska, L.; White, R.E.; Franz, M.; Ruhmann, M.; Allday, M.J. Epigenetic Repression of p16INK4A by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 2010, 6, e1000951. [Google Scholar] [CrossRef]
- Tsai, C.-L.; Li, H.-P.; Lu, Y.-J.; Hsueh, C.; Liang, Y.; Chen, C.-L.; Tsao, S.W.; Tse, K.-P.; Yu, J.-S.; Chang, Y.-S. Activation of DNA Methyltransferase 1 by EBV LMP1 Involves c-Jun NH 2 -Terminal Kinase Signaling. Cancer Res. 2006, 66, 11668–11676. [Google Scholar] [CrossRef]
- Dai, W.; Zheng, H.; Cheung, A.K.L.; Lung, M.L. Genetic and epigenetic landscape of nasopharyngeal carcinoma. Chinese Clin. Oncol. 2016, 5, 16. [Google Scholar] [CrossRef]
- García-Gutiérrez, L.; McKenna, S.; Kolch, W.; Matallanas, D. RASSF1A Tumour Suppressor: Target the Network for Effective Cancer Therapy. Cancers 2020, 12, 229. [Google Scholar] [CrossRef]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Li, Y.-Q.; Liu, N.; Sun, Y.; He, Q.-M.; Jiang, N.; Xu, Y.-F.; Chen, L.; Ma, J. 5-Azacytidine Enhances the Radiosensitivity of CNE2 and SUNE1 Cells In Vitro and In Vivo Possibly by Altering DNA Methylation. PLoS ONE 2014, 9, e93273. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.Y.; Deng, X.B.; Lin, X.T.; Jiang, L.L.; Huang, X.T.; Mo, Z.W.; Yuan, Y.W.; Teh, M.T. RASSF1A inhibits PDGFB-driven malignant phenotypes of nasopharyngeal carcinoma cells in a YAP1-dependent manner. Cell Death Dis. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
| Gene | Location | Roles | References |
|---|---|---|---|
| ADAMTS9 | 3p14.1 | Angiogenesis | [6] |
| PTPRG | 3p14.2 | Chromosomal translocations and deletions, cell cycle | [18] |
| BLU | 3p21.3 | Cell progress, stress-response | [7,16,17,19,20] |
| GNAT1 | 3p21.3 | Remains to be revealed | [21] |
| LARS2 | 3p21.3 | Protein synthesis | [22] |
| LTF | 3p21.3 | Immunomodulatory, homeostasis, anti-tumor activity, cell growth, cell cycle regulatory | [23,24,25] |
| MLH1 | 3p21.3 | Mismatch repair | [26,27] |
| RASSF1A | 3p21.3 | Cell proliferation, cell cycle regulation, apoptosis, micro-tubular stabilization | [5,13,26,27,28,29,30,31,32,33,34,35,36,37,38] |
| ITGA9 | 3p21.3 | Cell––cell and cell–matrix adhesion | [15] |
| DLEC1 | 3p22.2 | Cell communication, signaling transduction, cell proliferation | [7,38,39,40] |
| RAR-β | 3p24.2 | Hormone receptor, transcriptional regulator, retinoic acid signaling, cell growth and differentiation | [28,30,36] |
| VLH1 | 3p25.3 | Ubiquitination | [27] |
| TIG1 | 3q25.3 | Cell-to-cell contact | [39,41] |
| NPC Case (n) | Non-Cancerous Control (n) | Case | Control | Sp, Se, Po, Ne | References |
|---|---|---|---|---|---|
| Primary tumor tissue (28) | Nasopharyngeal epithelium (6) | 14/21 (66.67%) | 0/6 (0.00%) | Sp = 100.00% Se = 66.67% Po = 100.00% Ne = 46.15% | [34] |
| Primary tumor tissue (28) | Tissue (6) | 23/28 (82.14%) | 0/6 (0.00%) | Sp = 100.00% Se = 82.14% Po = 100.00% Ne = 54.55% | [30] |
| Nasopharyngeal brushing (28) | Nasopharyngeal brushing (12) | 11/28 (39.29%) | 0/26 (0.00%) | Sp = 100.00% Se = 39.29% Po = 100.00% Ne = 60.47% | [31] |
| Primary tumor tissue (30) | Tissue (6) | 20/30 (66.67%) | 0/6 (0.00%) | Sp = 100.00% Se = 66.67% Po = 100.00% Ne = 37.50% | [29] |
| Nasopharyngeal brushing (30) | Nasopharyngeal brushing (37) | 10/30 (33.33%) | 0/37 (0.00%) | Sp = 100.00% Se = 33.33% Po = 100.00% Ne = 64.91% | |
| M&T rinsing fluid (30) | M&T rinsing fluid (43) | 11/30 (36.67%) | 0/43 (0.00%) | Sp = 100.00% Se = 36.67% Po = 100.00% Ne = 69.36% | |
| Plasma (30) | Plasma (43) | 9/30 (30.00%) | 1/43 (2.32%) | Sp = 97.67% Se = 30.00% Po = 90.00% Ne = 66.67% | |
| Buffy coat (30) | Buffy coat (43) | 0/30 (0.00%) | 1/43 (2.32%) | Sp = 97.67% Se = 0.00% Po = 0.00% Ne = 58.33% | |
| Tumor tissue (28) | Tissue (5) | 13/28 (46.43%) | 0/5 (0.00%) | Sp = 100.00% Se = 46.43% Po = 100.00% Ne = 25.00% | [27] |
| Plasma (41) | Plasma (43) | 2/41 (4.88%) | 0/43 (0.00%) | Sp = 100.00% Se = 4.88% Po = 100.00% Ne = 52.44% | [26] |
| Tumor tissue (68) | Tissue (9) | 62/68 (91.18%) | 0/9 (0.00%) | Sp = 100.00% Se = 91.18% Po = 100.00% Ne = 60.00% | [36] |
| Tumor tissue (38) | Tissue (14) | 27/38 (71.05%) | 0/14 (0.00%) | Sp = 100.00% Se = 71.05% Po = 100.00% Ne = 56.00% | [33] |
| Tumor tissue/brushing (53) | Nasopharyngeal brushing (25) | 40/53 (75.47%) | 1/25 (4.00%) | Sp = 96.00% Se = 45.47% Po = 97.56% Ne = 64.87% | [5] |
| Tumor tissue (36) | Tissue (19) | 27/36 (75.00%) | 0/19 (0.00%) | Sp = 100.00% Se = 75.00% Po = 100.00% Ne = 67.86% | [28] |
| Tumor tissue (49) | Tissue (20) | 39/49 (79.59%) | 0/20 (0.00%) | Sp = 100.00% Se = 79.59% Po = 100.00% Ne = 66.67% | [32] |
| Nasopharyngeal brushing (49) | Nasopharyngeal brushing (20) | 29/49 (59.18%) | 0/20 (0.00%) | Sp = 100.00% Se = 59.18% Po = 100.00% Ne = 50.00% | |
| Serum (40) | Serum (41) | 7/40 (17.50%) | 2/41 (4.87%) | Sp = 95.12% Se = 17.50% Po = 77.78% Ne = 54.17% | [38] |
| Tumor tissue (70) | Nasopharyngeal brushing (70) | 47/70 (52.22%) | 9/70 (12.86%) | Sp = 87.14% Se = 67.14% Po = 83.93% Ne = 72.62% | [35] |
| Panel of Genes | NPC Case (n) | Non-Cancerous Control (n) | MI | Sp, Se, Po, Ne | References |
|---|---|---|---|---|---|
| RASSF1A, RAR-β, DAPK, p16, p15, p14, MGMT, GSTP1 | Primary tumor tissue (28) | Nasopharyngeal epithelium (6) | 28/28 (100.00%) of cases, at least one of seven genes: RASSF1A, RAR-β, DAPK, p16, p15, p14, and MGMT. All controls were not unmethylated. | Sp = 100.00% Se = 100.00% Po = 100.00% Ne = 100.00% | [30] |
| RASSF1A, DAPK, p16 | Nasopharyngeal brushing (28) | Nasopharyngeal brushing (12) | 22/28 (78.57%) of cases, at least one of three genes. All controls were not unmethylated. | Sp = 78.57% Se = 100.00% Po = 100.00% Ne = 66.67% | [31] |
| RASSF1A, E-cadherin, DAPK, p15, p16 | Primary tumor tissue (30) | Tissue (6) | 29/30 of cases, at least one of three genes. All controls were not unmethylated. | Sp = 100.00% Se = 96.97% Po = 100.00% Ne = 85.71% | [29] |
| Nasopharyngeal brushing (30) | Nasopharyngeal brushing (37) | 24/30 (80.00%) of cases, at least one of three genes. All controls were not unmethylated. | Sp = 100.00% Se = 80.00% Po = 100.00% Ne = 86.05% | ||
| M&T rinsing fluid (30) | M&T rinsing fluid (43) | 26/30 (87.00%) of cases, at least one of three genes. 1/43 (2.32%) of controls, at least one of three genes. | Sp = 97.67% Se = 86.67% Po = 96.30% Ne = 91.30% | ||
| Plasma (30) | Plasma (43) | 3/30 (10.00%) of cases, at least one of three genes. 2/43 (4.65%) of controls, at least one of three genes. | Sp = 95.34% Se = 10.00% Po = 60.00% Ne = 60.30% | ||
| Buffy coat (30) | Buffy coat (43) | 12/30 (40.00%) of cases, at least one of three genes. 3/43 (6.97%) of controls, at least one of three genes. | Sp = 93.02% Se = 40.00% Po = 80.00% Ne = 68.97% | ||
| RASSF1A, MLH1, CDH1, CDK2B, THBS1, MGMT, CDKN2A, TP73, C8, ARF, VHL | Tumor tissue (28) | Tissue (5) | 26/28 (92.86%) of cases, at least one of ten genes: RASSF1A, MLH1, CDH1, CDK2B, THBS1, MGMT, CDKN2A, TP73, C8, and ARF. All controls were not unmethylated. | Sp = 100.00% Se = 92.86% Po = 100.00% Ne = 71.43% | [27] |
| RASSF1A, MLH1, CDH1, DAPK, p15, p16 | Plasma (41) | Plasma (43) | 29/41 (70.73%) of cases, at least one of six genes. 4/43 (9.30%) of controls, at least one of six genes. | Sp = 90.70% Se = 70.73% Po = 87.88% Ne = 76.47% | [26] |
| RASSF1A, RAR-β, DAPK | Tumor tissue (68) | Tissue (9) | 67/68 (98.53%) of cases, at least one of three genes. All controls were not unmethylated. | Sp = 100.00% Se = 91.18% Po = 100.00% Ne = 60.00% | [36] |
| RASSF1A, CHFR, RIZ1, WIFI1, p16, RASSF2A, DAPK1, DLC1, CDH13, CADM1 | Tumor tissue/brushing (53) | Nasopharyngeal brushing (25) | 52/53 (98.11%) of cases, at least one of ten genes. nc in controls. | nc | [5] |
| RASSF1A, RAR-β, SHP1, DAPK, p16, GSTP1, TIMP3, APC, CDH1, MGMT | Tumor tissue (36) | Tissue (19) | nc | nc | [28] |
| RASSF1A, DAPK | Tumor tissue (49) | Tissue (20) | nc | nc | [32] |
| Nasopharyngeal brushing (49) | Nasopharyngeal brushing (20) | nc | nc | ||
| RASSF1A, DLEC1, CDKN2A, DAPK, UCHL1 | Serum (40) | Serum (41) | 34/40 (85.00%) of cases, at least one of five genes. 15/41 (36.59%) of controls, at least one of five genes. | Sp = 85.00% Se = 63.42% Po = 69.39% Ne = 81.25% | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lao, T.D.; Nguyen, T.N.; Le, T.A.H. Promoter Hypermethylation of Tumor Suppressor Genes Located on Short Arm of the Chromosome 3 as Potential Biomarker for the Diagnosis of Nasopharyngeal Carcinoma. Diagnostics 2021, 11, 1404. https://doi.org/10.3390/diagnostics11081404
Lao TD, Nguyen TN, Le TAH. Promoter Hypermethylation of Tumor Suppressor Genes Located on Short Arm of the Chromosome 3 as Potential Biomarker for the Diagnosis of Nasopharyngeal Carcinoma. Diagnostics. 2021; 11(8):1404. https://doi.org/10.3390/diagnostics11081404
Chicago/Turabian StyleLao, Thuan Duc, Toan Ngoc Nguyen, and Thuy Ai Huyen Le. 2021. "Promoter Hypermethylation of Tumor Suppressor Genes Located on Short Arm of the Chromosome 3 as Potential Biomarker for the Diagnosis of Nasopharyngeal Carcinoma" Diagnostics 11, no. 8: 1404. https://doi.org/10.3390/diagnostics11081404
APA StyleLao, T. D., Nguyen, T. N., & Le, T. A. H. (2021). Promoter Hypermethylation of Tumor Suppressor Genes Located on Short Arm of the Chromosome 3 as Potential Biomarker for the Diagnosis of Nasopharyngeal Carcinoma. Diagnostics, 11(8), 1404. https://doi.org/10.3390/diagnostics11081404






