A Comprehensive Review on Radiomics and Deep Learning for Nasopharyngeal Carcinoma Imaging
Abstract
:1. Introduction
- The pipeline of radiomics and the principle of DL are briefly described;
- The studies of radiomics and DL for NPC imaging in recent years are summarized;
- The deficiencies of current studies and the potential of radiomics and DL for NPC imaging are discussed.
2. Pipeline of Radiomics
3. The Principle of DL
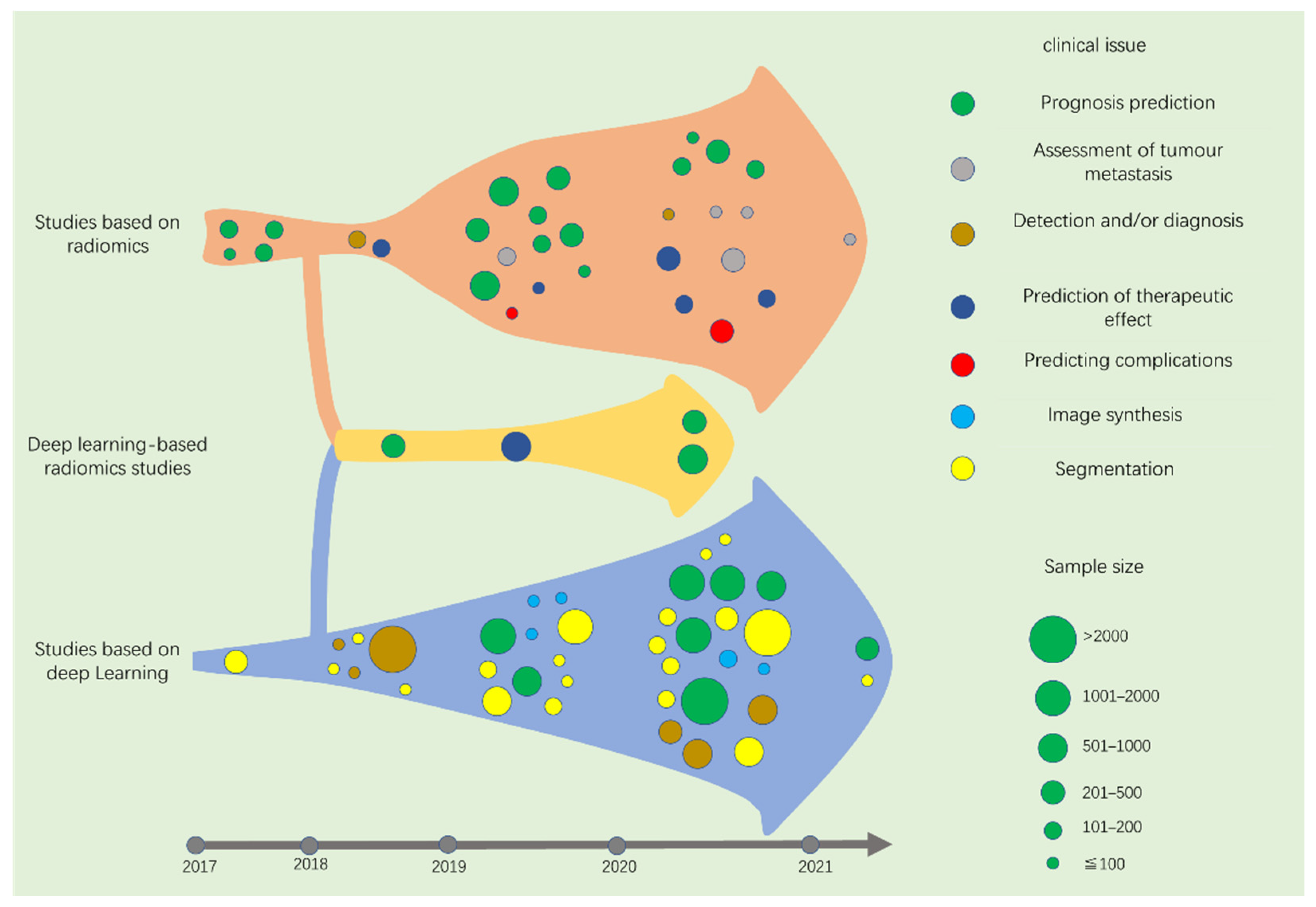
4. Screening of Studies
5. Studies Based on Radiomics
5.1. Prognosis Prediction
5.1.1. 2017
5.1.2. 2018
5.1.3. 2019
5.1.4. 2020
5.1.5. 2021
5.2. Assessment of Tumour Metastasis
5.2.1. 2017–2018
5.2.2. 2019
5.2.3. 2020
5.2.4. 2021
5.3. Tumour Diagnosis
5.3.1. 2017
5.3.2. 2018
5.3.3. 2019
5.3.4. 2020
5.4. Prediction of Therapeutic Effect
5.4.1. 2017
5.4.2. 2018
5.4.3. 2019
5.4.4. 2020
5.5. Predicting Complications
5.5.1. 2017–2018
5.5.2. 2019
5.5.3. 2020
6. Studies Based on DL
6.1. Prognosis Prediction
6.1.1. 2017–2018
6.1.2. 2019
6.1.3. 2020
6.1.4. 2021
6.2. Image Synthesis
6.2.1. 2017–2018
6.2.2. 2019
6.2.3. 2020
6.3. Detection and/or Diagnosis
6.3.1. 2017
6.3.2. 2018
6.3.3. 2019
6.3.4. 2020
6.4. Segmentation
6.4.1. 2017
6.4.2. 2018
6.4.3. 2019
6.4.4. 2020
6.4.5. 2021
7. Deep Learning-Based Radiomics
7.1. Studies Based on Deep Learning-Based Radiomics (DLR)
7.1.1. 2017
7.1.2. 2018
7.1.3. 2019
7.1.4. 2020
8. Discussion
9. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/today (accessed on 31 March 2021).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.M.; Hussain, S.S.M. Incidence of nasopharyngeal carcinoma in Chinese immigrants, compared with Chinese in China and South East Asia: Review. J. Laryngol. Otol. 2009, 123, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Bei, J.X.; Su, W.H.; Ng, C.C.; Yu, K.; Chin, Y.M.; Lou, P.J.; Hsu, W.L.; McKay, J.D.; Chen, C.J.; Chang, Y.S.; et al. A GWAS meta-analysis and replication study identifies a novel locus within CLPTM1L/TERT associated with Nasopharyngeal carcinoma in individuals of Chinese ancestry. Cancer Epidemiol. Prev. Biomark. 2016, 25, 188–192. [Google Scholar] [CrossRef] [Green Version]
- Cui, Q.; Feng, Q.-S.; Mo, H.-Y.; Sun, J.; Xia, Y.-F.; Zhang, H.; Foo, J.N.; Guo, Y.-M.; Chen, L.-Z.; Li, M.; et al. An extended genome-wide association study identifies novel susceptibility loci for nasopharyngeal carcinoma. Hum. Mol. Genet. 2016, 25, 3626–3634. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.A.; Woo, J.K.; King, A.; Zee, B.C.-Y.; Lam, W.K.J.; Chan, S.; Chu, S.W.; Mak, C.; Tse, I.O.; Leung, S.Y.; et al. Analysis of Plasma Epstein–Barr Virus DNA to Screen for Nasopharyngeal Cancer. N. Engl. J. Med. 2017, 377, 513–522. [Google Scholar] [CrossRef]
- Eveson, J.W.; Auclair, P.; Gnepp, D.R.; El-Naggar, A.K.; Barnes, L. World Health Organization Classification of Tumours: Pathology and Genetics of Head and Neck Tumours; Barnes, L., Eveson, J.W., Reichart, P., Sidransky, D., Eds.; IARC Press: Lyon, France, 2005; pp. 88–111. [Google Scholar]
- Lee, A.W.; Ng, W.T.; Chan, L.L.; Hung, W.M.; Chan, C.C.; Sze, H.C.; Chan, O.S.H.; Chang, A.T.; Yeung, R.M. Evolution of treatment for nasopharyngeal cancer—Success and setback in the intensity-modulated radiotherapy era. Radiother. Oncol. 2014, 110, 377–384. [Google Scholar] [CrossRef]
- Butterfield, D. Impacts of Water and Export Market Restrictions on Palestinian Agriculture. Toronto: McMaster University and Econometric Research Limited, Applied Research Institute of Jerusalem (ARIJ). Available online: http://www.socserv.mcmaster.ca/kubursi/ebooks/water.htm (accessed on 31 March 2021).
- Chua, M.L.K.; Wee, J.T.S.; Hui, E.P.; Chan, A.T.C. Nasopharyngeal carcinoma. Lancet 2016, 387, 1012–1024. [Google Scholar] [CrossRef]
- Yuan, H.; Lai-Wan, K.D.; Kwong, D.L.-W.; Fong, D.; King, A.; Vardhanabhuti, V.; Lee, V.; Khong, P.-L. Cervical nodal volume for prognostication and risk stratification of patients with nasopharyngeal carcinoma, and implications on the TNM-staging system. Sci. Rep. 2017, 7, 10387. [Google Scholar] [CrossRef] [Green Version]
- Chan, A.T.C.; Grégoire, V.; Lefebvre, J.L.; Licitra, L.; Hui, E.P.; Leung, S.F.; Felip, E. Nasopharyngeal cancer: EHNS-ESMO-ESTRO clinical practice guidelines for diagnosis, treatment andfollow-up. Ann. Oncol. 2012, 23, vii83–vii85. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Sham, J.S. Nasopharyngeal carcinoma. Lancet 2005, 365, 2041–2054. [Google Scholar] [CrossRef]
- Vokes, E.E.; Liebowitz, D.N.; Weichselbaum, R.R. Nasopharyngeal carcinoma. Lancet 1997, 350, 1087–1091. [Google Scholar] [CrossRef]
- Pohlhaus, J.R.; Cook-Deegan, R.M. Genomics Research: World Survey of Public Funding. BMC Genom. 2008, 9, 472. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Data from: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [Green Version]
- Rahmim, A.; Schmidtlein, C.R.; Jackson, A.; Sheikhbahaei, S.; Marcus, C.; Ashrafinia, S.; Soltani, M.; Subramaniam, R.M. A novel metric for quantification of homogeneous and heterogeneous tumors in PET for enhanced clinical outcome prediction. Phys. Med. Biol. 2015, 61, 227–242. [Google Scholar] [CrossRef] [Green Version]
- Buvat, I.; Orlhac, F.; Soussan, M.; Orhlac, F. Tumor Texture Analysis in PET: Where Do We Stand? J. Nucl. Med. 2015, 56, 1642–1644. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Tian, J.; Dong, D.; Gu, D.; Dong, Y.; Zhang, L.; Lian, Z.; Liu, J.; Luo, X.; Pei, S.; et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin. Cancer Res. 2017, 23, 4259–4269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.; Oikonomou, A.; Benali, H. From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and Opportunities. IEEE Signal Process. Mag. 2019, 36, 132–160. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics 2019, 9, 1303–1322. [Google Scholar] [CrossRef]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef]
- Ford, J.; Dogan, N.; Young, L.; Yang, F. Quantitative Radiomics: Impact of Pulse Sequence Parameter Selection on MRI-Based Textural Features of the Brain. Contrast Media Mol. Imaging 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Dong, D.; Fang, M.-J.; Li, L.; Tang, L.-L.; Chen, L.; Li, W.-F.; Mao, Y.-P.; Fan, W.; Liu, L.-Z.; et al. Prognostic Value of Deep Learning PET/CT-Based Radiomics: Potential Role for Future Individual Induction Chemotherapy in Advanced Nasopharyngeal Carcinoma. Clin. Cancer Res. 2019, 25, 4271–4279. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Hu, Y.; Qiao, M.; Wang, Y.; Yu, J.; Li, J.; Chang, C. Radiomics Analysis on Ultrasound for Prediction of Biologic Behavior in Breast Invasive Ductal Carcinoma. Clin. Breast Cancer 2018, 18, e335–e344. [Google Scholar] [CrossRef]
- Song, G.; Xue, F.; Zhang, C. A Model Using Texture Features to Differentiate the Nature of Thyroid Nodules on Sonography. J. Ultrasound Med. 2015, 34, 1753–1760. [Google Scholar] [CrossRef]
- Polan, D.F.; Brady, S.L.; Kaufman, R.A. Tissue segmentation of computed tomography images using a Random Forest algorithm: A feasibility study. Phys. Med. Biol. 2016, 61, 6553. [Google Scholar] [CrossRef] [PubMed]
- Qiang, M.; Li, C.; Sun, Y.; Sun, Y.; Ke, L.; Xie, C.; Zhang, T.; Zou, Y.; Qiu, W.; Gao, M.; et al. A Prognostic Predictive System Based on Deep Learning for Locoregionally Advanced Nasopharyngeal Carcinoma. J. Natl. Cancer Inst. 2021, 113, 606–615. [Google Scholar] [CrossRef]
- Aerts, H.J. Data Science in Radiology: A Path Forward. Clin. Cancer Res. 2017, 24, 532–534. [Google Scholar] [CrossRef] [Green Version]
- Hatt, M.; Tixier, F.; Pierce, L.; Kinahan, P.; Le Rest, C.C.; Visvikis, D. Characterization of PET/CT images using texture analysis: The past, the present… any future? Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 151–165. [Google Scholar] [CrossRef]
- Zhang, L.; Fried, D.V.; Fave, X.J.; Hunter, L.A.; Yang, J.; Court, L.E. IBEX: An open infrastructure software platform to facilitate collaborative work in radiomics. Med. Phys. 2015, 42, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Bagherzadeh-Khiabani, F.; Ramezankhani, A.; Azizi, F.; Hadaegh, F.; Steyerberg, E.W.; Khalili, D. A tutorial on variable selection for clinical prediction models: Feature selection methods in data mining could improve the results. J. Clin. Epidemiol. 2016, 71, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, Z.; Shen, C.; Li, Z.; Yan, F.; Tian, J.; Xian, J. MR-based radiomics signature in differentiating ocular adnexal lymphoma from idiopathic orbital inflammation. Eur. Radiol. 2018, 28, 3872–3881. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.E.; Jo, Y.; Shim, W.H.; Nam, S.J.; Kim, J.H.; Yoo, R.E.; Choi, S.H.; Kim, H.S. Incorporating diffusion-and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro-Oncology 2019, 21, 404–414. [Google Scholar] [CrossRef]
- Zhang, Y.; Oikonomou, A.; Wong, A.; Haider, M.A.; Khalvati, F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, srep46349. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, W.; Tixier, F.; van der Vos, C.S.; Vriens, D.; Le Rest, C.C.; Bussink, J.; Oyen, W.J.; de Geus-Oei, L.-F.; Visvikis, D.; Visser, E.P. The Impact of Optimal Respiratory Gating and Image Noise on Evaluation of Intratumor Heterogeneity on 18F-FDG PET Imaging of Lung Cancer. J. Nucl. Med. 2016, 57, 1692–1698. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, D.; Poellinger, A.; Shao, L.; Reyes, M. Interpretability-Driven Sample Selection Using Self Supervised Learning for Disease Classification and Segmentation. IEEE Trans. Med. Imaging 2021. [Google Scholar] [CrossRef]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in breast cancer classification and prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef]
- Hawkins, S.; Wang, H.; Liu, Y.; Garcia, A.; Stringfield, O.; Krewer, H.; Li, Q.; Cherezov, D.; Gatenby, R.A.; Balagurunathan, Y.; et al. Predicting Malignant Nodules from Screening CT Scans. J. Thorac. Oncol. 2016, 11, 2120–2128. [Google Scholar] [CrossRef] [Green Version]
- Verduin, M.; Primakov, S.; Compter, I.; Woodruff, H.; van Kuijk, S.; Ramaekers, B.; Dorsthorst, M.T.; Revenich, E.; ter Laan, M.; Pegge, S.; et al. Prognostic and Predictive Value of Integrated Qualitative and Quantitative Magnetic Resonance Imaging Analysis in Glioblastoma. Cancers 2021, 13, 722. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, H.; Wu, J.; Chen, C.; Yuan, Q.; Huang, W.; Li, T.; Xi, S.; Hu, Y.; Zhou, Z.; et al. Noninvasive imaging evaluation of tumor immune microenvironment to predict outcomes in gastric cancer. Ann. Oncol. 2020, 31, 760–768. [Google Scholar] [CrossRef]
- Lian, C.; Ruan, S.; Denœux, T.; Jardin, F.; Vera, P. Selecting radiomic features from FDG-PET images for cancer treatment outcome prediction. Med. Image Anal. 2016, 32, 257–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Cao, J.; Hu, J.; Topatana, W.; Li, S.; Juengpanich, S.; Lin, J.; Tong, C.; Shen, J.; Zhang, B.; et al. Clinical-Radiomic Analysis for Pretreatment Prediction of Objective Response to First Transarterial Chemoembolization in Hepatocellular Carcinoma. Liver Cancer 2021, 10, 38–51. [Google Scholar] [CrossRef]
- Carles, M.; Fechter, T.; Radicioni, G.; Schimek-Jasch, T.; Adebahr, S.; Zamboglou, C.; Nicolay, N.; Martí-Bonmatí, L.; Nestle, U.; Grosu, A.; et al. FDG-PET Radiomics for Response Monitoring in Non-Small-Cell Lung Cancer Treated with Radiation Therapy. Cancers 2021, 13, 814. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zheng, J.; Shen, J.; Yuan, Z.; Xie, M.; Gao, M.; Tan, H.; Liang, Z.-G.; Rong, X.; Li, Y.; et al. A Radiomics Model for Predicting the Response to Bevacizumab in Brain Necrosis after Radiotherapy. Clin. Cancer Res. 2020, 26, 5438–5447. [Google Scholar] [CrossRef] [PubMed]
- Nie, K.; Shi, L.; Chen, Q.; Hu, X.; Jabbour, S.K.; Yue, N.; Niu, T.; Sun, X. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clin. Cancer Res. 2016, 22, 5256–5264. [Google Scholar] [CrossRef] [Green Version]
- Samiei, S.; Granzier, R.; Ibrahim, A.; Primakov, S.; Lobbes, M.; Beets-Tan, R.; van Nijnatten, T.; Engelen, S.; Woodruff, H.; Smidt, M. Dedicated Axillary MRI-Based Radiomics Analysis for the Prediction of Axillary Lymph Node Metastasis in Breast Cancer. Cancers 2021, 13, 757. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Deng, Y.; Liu, T.; Zhou, J.; Jia, X.; Xiao, T.; Zhou, S.; Li, J.; Guo, Y.; Wang, Y.; et al. Lymph node metastasis prediction of papillary thyroid carcinoma based on transfer learning radiomics. Nat. Commun. 2020, 11, 4807. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Q.; Zhang, C.; Sun, J.; He, K.; Xie, Y.; Zhang, Y.; Fu, Y.; Zhang, H. Multiregional-Based Magnetic Resonance Imaging Radiomics Combined with Clinical Data Improves Efficacy in Predicting Lymph Node Metastasis of Rectal Cancer. Front. Oncol. 2021, 10, 10. [Google Scholar] [CrossRef]
- Mouraviev, A.; Detsky, J.; Sahgal, A.; Ruschin, M.; Lee, Y.K.; Karam, I.; Heyn, C.; Stanisz, G.J.; Martel, A.L. Use of radiomics for the prediction of local control of brain metastases after stereotactic radiosurgery. Neuro-Oncology 2020, 22, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yuan, Q.; Lv, W.; Xi, S.; Huang, W.; Sun, Z.; Chen, H.; Zhao, L.; Liu, W.; Hu, Y.; et al. Radiomic signature of 18F fluorodeoxyglucose PET/CT for prediction of gastric cancer survival and chemotherapeutic benefits. Theranostics 2018, 8, 5915–5928. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.K.; Júnior, J.R.F.; Wada, D.T.; Tenório, A.P.M.; Barbosa, M.H.N.; Marques, P.M.D.A. Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: Advances in imaging towards to precision medicine. Radiol. Bras. 2019, 52, 387–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef]
- Moor, J. The Dartmouth College artificial intelligence conference: The next fifty years. Ai Mag. 2006, 27, 87. [Google Scholar]
- Zhou, X.; Li, C.; Rahaman, M.; Yao, Y.; Ai, S.; Sun, C.; Wang, Q.; Zhang, Y.; Li, M.; Li, X.; et al. A Comprehensive Review for Breast Histopathology Image Analysis Using Classical and Deep Neural Networks. IEEE Access 2020, 8, 90931–90956. [Google Scholar] [CrossRef]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Foster, K.R.; Koprowski, R.; Skufca, J.D. Machine learning, medical diagnosis, and biomedical engineering research—Commentary. Biomed. Eng. Online 2014, 13, 94. [Google Scholar] [CrossRef] [Green Version]
- Rashidi, H.H.; Tran, N.K.; Betts, E.V.; Howell, L.P.; Green, R. Artificial intelligence and machine learning in pathology: The present landscape of supervised methods. Acad. Pathol. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Wang, Y.; Amiryousefi, A.; Tang, J. Unsupervised Learning and Multipartite Network Models: A Promising Approach for Understanding Traditional Medicine. Front. Pharmacol. 2020, 11, 1319. [Google Scholar] [CrossRef]
- Bzdok, D.; Krzywinski, M.; Altman, N. Machine learning: Supervised methods. Nat. Methods 2018, 15, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Zipser, D.; Andersen, R.A. A back-propagation programmed network that simulates response properties of a subset of posterior parietal neurons. Nat. Cell Biol. 1988, 331, 679–684. [Google Scholar] [CrossRef]
- Kriegeskorte, N.; Golan, T. Neural network models and deep learning. Curr. Biol. 2019, 29, R231–R236. [Google Scholar] [CrossRef] [PubMed]
- Manisha; Dhull, S.K.; Singh, K.K. ECG Beat Classifiers: A Journey from ANN To DNN. Procedia Comput. Sci. 2020, 167, 747–759. [Google Scholar] [CrossRef]
- Soffer, S.; Ben-Cohen, A.; Shimon, O.; Amitai, M.M.; Greenspan, H.; Klang, E. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology 2019, 290, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Bao, F.; Dai, Q.; Wu, L.F.; Altschuler, S.J. Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. Nat. Methods 2019, 16, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, X.; Zhou, X.; Cheng, P.; He, K.; Li, C. Knowledge Enhanced LSTM for Coreference Resolution on Biomedical Texts. Bioinformatics 2021. [Google Scholar] [CrossRef]
- Hua, Y.; Guo, J.; Zhao, H. Deep belief networks and deep learning. In Proceedings of the 2015 International Conference on Intelligent Computing and Internet of Things, Harbin, China, 17–18 January 2015; pp. 1–4. [Google Scholar]
- Ko, W.Y.; Siontis, K.C.; Attia, Z.I.; Carter, R.E.; Kapa, S.; Ommen, S.R.; Demuth, S.J.; Ackerman, M.J.; Gersh, B.J.; Arruda-Olson, A.M.; et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J. Am. Coll. Cardiol. 2020, 75, 722–733. [Google Scholar] [CrossRef]
- Skrede, O.-J.; De Raedt, S.; Kleppe, A.; Hveem, T.S.; Liestøl, K.; Maddison, J.; Askautrud, H.; Pradhan, M.; Nesheim, J.A.; Albregtsen, F.; et al. Deep learning for prediction of colorectal cancer outcome: A discovery and validation study. Lancet 2020, 395, 350–360. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; 773p. [Google Scholar]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Rome, Italy, 26–28 July 2019; pp. 6105–6114. [Google Scholar]
- Zhang, B.; He, X.; Ouyang, F.; Gu, D.; Dong, Y.; Zhang, L.; Mo, X.; Huang, W.; Tian, J.; Zhang, S. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017, 403, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ouyang, F.; Gu, D.; Dongsheng, G.; Zhang, L.; Mo, X.; Huang, W.; Zhang, S. Advanced nasopharyngeal carcinoma: Pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget 2017, 8, 72457–72465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, F.S.; Guo, B.L.; Zhang, B.; Dong, Y.H.; Zhang, L.; Mo, X.K.; Huang, W.H.; Zhang, S.X.; Hu, Q.G. Exploration and validation of radiomics signature as an independent prognostic biomarker in stage III-IVb Nasopharyngeal carcinoma. Oncotarget 2017, 8, 74869–74879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, W.; Yuan, Q.; Wang, Q.; Ma, J.; Feng, Q.; Chen, W.; Rahmim, A.; Lu, L. Radiomics Analysis of PET and CT Components of PET/CT Imaging Integrated with Clinical Parameters: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2019, 21, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, E.-H.; Zhang, W.-J.; Li, H.-J.; Zhang, G.-Y.; Jing, B.-Z.; Zhou, J.; Cui, C.-Y.; Chen, M.-Y.; Sun, Y.; Liu, L.-Z.; et al. Radiomics on multi-modalities MR sequences can subtype patients with non-metastatic nasopharyngeal carcinoma (NPC) into distinct survival subgroups. Eur. Radiol. 2019, 29, 5590–5599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Huang, M.Y.; Li, Y.; Liang, J.H.; Gao, T.S.; Deng, B.; Yao, J.J.; Lin, L.; Chen, F.P.; Huang, X.D.; et al. Pretreatment MRI radiomics analysis allows for reliable prediction of local recurrence in non-metastatic T4 Nasopharyngeal carcinoma. EBioMedicine 2019, 42, 270–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Tian, J.; Zhang, B.; Li, M.; Xie, W.; Zou, Y.; Tan, Q.; Liu, L.; Zhu, J.; Shou, A.; et al. A multidimensional nomogram combining overall stage, dose volume histogram parameters and radiomics to predict progression-free survival in patients with locoregionally advanced nasopharyngeal carcinoma. Oral Oncol. 2019, 98, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Oei, R.W.; Zhai, R.; Kong, F.; Du, C.; Hu, C.; Hu, W.; Zhang, Z.; Ying, H.; Wang, J. MRI-based radiomics signature is a quantitative prognostic biomarker for nasopharyngeal carcinoma. Sci. Rep. 2019, 9, 10412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhou, H.; Gu, D.; Tian, J.; Zhang, B.; Dong, D.; Mo, X.; Liu, J.; Luo, X.; Pei, S.; et al. Radiomic Nomogram: Pretreatment Evaluation of Local Recurrence in Nasopharyngeal Carcinoma based on MR Imaging. J. Cancer 2019, 10, 4217–4225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, J.; Fang, J.; Duan, X.; Yang, Z.; Cao, M.; Zhang, F.; Lu, L.; Zhang, X.; Wu, X.; Ding, Y.; et al. Predictive value of pretreatment MRI texture analysis in patients with primary nasopharyngeal carcinoma. Eur. Radiol. 2019, 29, 4105–4113. [Google Scholar] [CrossRef] [Green Version]
- Du, R.; Lee, V.H.; Yuan, H.; Lam, K.-O.; Pang, H.H.; Chen, Y.; Lam, E.Y.; Khong, P.-L.; Lee, A.W.; Kwong, D.L.; et al. Radiomics Model to Predict Early Progression of Nonmetastatic Nasopharyngeal Carcinoma after Intensity Modulation Radiation Therapy: A Multicenter Study. Radiol. Artif. Intell. 2019, 1, e180075. [Google Scholar] [CrossRef]
- Xu, H.; Lv, W.; Feng, H.; Du, D.; Yuan, Q.; Wang, Q.; Dai, Z.; Yang, W.; Feng, Q.; Ma, J.; et al. Subregional Radiomics Analysis of PET/CT Imaging with Intratumor Partitioning: Application to Prognosis for Nasopharyngeal Carcinoma. Mol. Imaging Biol. 2020, 22, 1414–1426. [Google Scholar] [CrossRef]
- Shen, H.; Wang, Y.; Liu, D.; Lv, R.; Huang, Y.; Peng, C.; Jiang, S.; Wang, Y.; He, Y.; Lan, X.; et al. Predicting Progression-Free Survival Using MRI-Based Radiomics for Patients with Nonmetastatic Nasopharyngeal Carcinoma. Front. Oncol. 2020, 10, 618. [Google Scholar] [CrossRef]
- Bologna, M.; Corino, V.; Calareso, G.; Tenconi, C.; Alfieri, S.; Iacovelli, N.A.; Cavallo, A.; Cavalieri, S.; Locati, L.; Bossi, P.; et al. Baseline MRI-Radiomics Can Predict Overall Survival in Non-Endemic EBV-Related Nasopharyngeal Carcinoma Patients. Cancers 2020, 12, 2958. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Liang, J.; Wang, L.; Niu, J.; Ge, X.; Pang, P.; Ding, Z. Radiomics Analysis and Correlation with Metabolic Parameters in Nasopharyngeal Carcinoma Based on PET/MR Imaging. Front. Oncol. 2020, 10, 1619. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Hong, X.; Yuan, Q.; Lu, L.; Wang, Q.; Chen, W. Prediction of local recurrence and distant metastasis using radiomics analysis of pretreatment nasopharyngeal [18F]FDG PET/CT images. Ann. Nucl. Med. 2021, 35, 458–468. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, D.; Li, H.; Tian, J.; Ouyang, F.; Mo, X.; Zhang, B.; Luo, X.; Lian, Z.; Pei, S.; et al. Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: A retrospective cohort study. EBioMedicine 2019, 40, 327–335. [Google Scholar] [CrossRef]
- Zhong, X.; Li, L.; Jiang, H.; Yin, J.; Lu, B.; Han, W.; Li, J.; Zhang, J. Cervical spine osteoradionecrosis or bone metastasis after radiotherapy for nasopharyngeal carcinoma? The MRI-based radiomics for characterization. BMC Med. Imaging 2020, 20, 104. [Google Scholar] [CrossRef]
- Akram, F.; Koh, P.E.; Wang, F.; Zhou, S.; Tan, S.H.; Paknezhad, M.; Park, S.; Hennedige, T.; Thng, C.H.; Lee, H.K.; et al. Exploring MRI based radiomics analysis of intratumoral spatial heterogeneity in locally advanced nasopharyngeal carcinoma treated with intensity modulated radiotherapy. PLoS ONE 2020, 15, e0240043. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, L.; Zhang, B.; Zhang, L.; Du, H.; Lu, L.; Zhang, S.; Yang, W.; Feng, Q. The effects of volume of interest delineation on MRI-based radiomics analysis: Evaluation with two disease groups. Cancer Imaging 2019, 19, 89–112. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Yuan, Q.; Wang, Q.; Ma, J.; Jiang, J.; Yang, W.; Feng, Q.; Chen, W.; Rahmim, A.; Lu, L. Robustness versus disease differentiation when varying parameter settings in radiomics features: Application to nasopharyngeal PET/CT. Eur. Radiol. 2018, 28, 3245–3254. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Feng, H.; Lv, W.; Ashrafinia, S.; Yuan, Q.; Wang, Q.; Yang, W.; Feng, Q.; Chen, W.; Rahmim, A.; et al. Machine Learning Methods for Optimal Radiomics-Based Differentiation Between Recurrence and Inflammation: Application to Nasopharyngeal Carcinoma Post-therapy PET/CT Images. Mol. Imaging Biol. 2020, 22, 730–738. [Google Scholar] [CrossRef]
- Wang, G.; He, L.; Yuan, C.; Huang, Y.; Liu, Z.; Liang, C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur. J. Radiol. 2018, 98, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-T.; Lam, S.-K.; To, L.-H.; Tse, K.-Y.; Cheng, N.-Y.; Fan, Y.-N.; Lo, C.-L.; Or, K.-W.; Chan, M.-L.; Hui, K.-C.; et al. Pretreatment Prediction of Adaptive Radiation Therapy Eligibility Using MRI-Based Radiomics for Advanced Nasopharyngeal Carcinoma Patients. Front. Oncol. 2019, 9, 1050. [Google Scholar] [CrossRef]
- Yongfeng, P.; Chuner, J.; Lei, W.; Fengqin, Y.; Zhimin, Y.; Zhenfu, F.; Haitao, J.; Yangming, J.; Fangzheng, W. The usefulness of pre-treatment MR-based radiomics on early response of neoadjuvant chemotherapy in patients with locally advanced Nasopharyngeal carcinoma. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2021, 28, 605–613. [Google Scholar]
- Zhang, L.; Ye, Z.; Ruan, L.; Jiang, M. Pretreatment MRI-Derived Radiomics May Evaluate the Response of Different Induction Chemotherapy Regimens in Locally advanced Nasopharyngeal Carcinoma. Acad. Radiol. 2020, 27, 1655–1664. [Google Scholar] [CrossRef]
- Zhao, L.; Gong, J.; Xi, Y.; Xu, M.; Li, C.; Kang, X.; Yin, Y.; Qin, W.; Yin, H.; Shi, M. MRI-based radiomics nomogram may predict the response to induction chemotherapy and survival in locally advanced nasopharyngeal carcinoma. Eur. Radiol. 2020, 30, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, H.; Huang, S.; Chen, X.; Zhou, H.; Chang, H.; Xia, Y.; Wang, G.; Yang, X. Early prediction of acute xerostomia during radiation therapy for nasopharyngeal cancer based on delta radiomics from CT images. Quant. Imaging Med. Surg. 2019, 9, 1288–1302. [Google Scholar] [CrossRef]
- Zhang, B.; Lian, Z.; Zhong, L.; Zhang, X.; Dong, Y.; Chen, Q.; Zhang, L.; Mo, X.; Huang, W.; Yang, W.; et al. Machine-learning based MRI radiomics models for early detection of radiation-induced brain injury in nasopharyngeal carcinoma. BMC Cancer 2020, 20, 502. [Google Scholar] [CrossRef] [PubMed]
- Qiang, M.; Lv, X.; Li, C.; Liu, K.; Chen, X.; Guo, X. Deep learning in nasopharyngeal carcinoma: A retrospective cohort study of 3D convolutional neural networks on magnetic resonance imaging. Ann. Oncol. 2019, 30, v471. [Google Scholar] [CrossRef]
- Du, R.; Cao, P.; Han, L.; Ai, Q.; King, A.D.; Vardhanabhuti, V. Deep convolution neural network model for automatic risk assessment of patients with non-metastatic Nasopharyngeal carcinoma. arXiv 2019, arXiv:1907.11861. [Google Scholar]
- Yang, Q.; Guo, Y.; Ou, X.; Wang, J.; Hu, C. Automatic T Staging Using Weakly Supervised Deep Learning for Nasopharyngeal Carcinoma on MR Images. J. Magn. Reson. Imaging 2020, 52, 1074–1082. [Google Scholar] [CrossRef]
- Jing, B.; Deng, Y.; Zhang, T.; Hou, D.; Li, B.; Qiang, M.; Liu, K.; Ke, L.; Li, T.; Sun, Y.; et al. Deep learning for risk prediction in patients with nasopharyngeal carcinoma using multi-parametric MRIs. Comput. Methods Programs Biomed. 2020, 197, 105684. [Google Scholar] [CrossRef]
- Cui, C.; Wang, S.; Zhou, J.; Dong, A.; Xie, F.; Li, H.; Liu, L. Machine Learning Analysis of Image Data Based on Detailed MR Image Reports for Nasopharyngeal Carcinoma Prognosis. BioMed Res. Int. 2020, 2020. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Xia, W.; Qiang, M.; Chen, X.; Liu, J.; Guo, X.; Lv, X. Deep learning pathological microscopic features in endemic nasopharyngeal cancer: Prognostic value and protentional role for individual induction chemotherapy. Cancer Med. 2019, 9, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, X.; Liu, J.; Zhang, B.; Mo, X.; Chen, Q.; Fang, J.; Wang, F.; Li, M.; Chen, Z.; et al. MRI-based deep-learning model for distant metastasis-free survival in locoregionally advanced Nasopharyngeal carcinoma. J. Magn. Reson. Imaging 2021, 53, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, J.; Liu, Z.; Teng, J.; Xie, Q.; Zhang, L.; Liu, X.; Shi, J.; Chen, L. A preliminary study of using a deep convolution neural network to generate synthesized CT images based on CBCT for adaptive radiotherapy of nasopharyngeal carcinoma. Phys. Med. Biol. 2019, 64, 145010. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Zhang, X.; Deng, W. Synthetic CT Generation Based on T2 Weighted MRI of Nasopharyngeal Carcinoma (NPC) Using a Deep Convolutional Neural Network (DCNN). Front. Oncol. 2019, 9, 1333. [Google Scholar] [CrossRef]
- Tie, X.; Lam, S.K.; Zhang, Y.; Lee, K.H.; Au, K.H.; Cai, J. Pseudo-CT generation from multi-parametric MRI using a novel multi-channel multi-path conditional generative adversarial network for Nasopharyngeal carcinoma patients. Med. Phys. 2020, 47, 1750–1762. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, S.; Qin, A.; Chen, M.; Gao, X.; Liu, Y.; Miao, J.; Gu, H.; Zhao, C.; Deng, X.; et al. Magnetic resonance-based synthetic computed tomography images generated using generative adversarial networks for nasopharyngeal carcinoma radiotherapy treatment planning. Radiother. Oncol. 2020, 150, 217–224. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abd Ghani, M.K.; Arunkumar, N.; Raed, H.; Mohamad, A.; Mohd, B. A real time computer aided object detection of Nasopharyngeal carcinoma using genetic algorithm and artificial neural network based on Haar feature fear. Future Gener. Comput Syst. 2018, 89, 539–547. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abd Ghani, M.K.; Arunkumar, N.; Hamed, R.I.; Mostafa, S.A.; Abdullah, M.K.; Burhanuddin, M.A. Decision support system for Nasopharyngeal carcinoma discrimination from endoscopic images using artificial neural network. J. Supercomput. 2020, 76, 1086–1104. [Google Scholar] [CrossRef]
- Abd Ghani, M.K.; Mohammed, M.A.; Arunkumar, N.; Mostafa, S.; Ibrahim, D.A.; Abdullah, M.K.; Jaber, M.M.; Abdulhay, E.; Ramirez-Gonzalez, G.; Burhanuddin, M.A. Decision-level fusion scheme for Nasopharyngeal carcinoma identification using machine learning techniques. Neu. Comput. Appl. 2020, 32, 625–638. [Google Scholar] [CrossRef]
- Li, C.; Jing, B.; Ke, L.; Li, B.; Xia, W.; He, C.; Qian, C.; Zhao, C.; Mai, H.; Chen, M.; et al. Development and validation of an endoscopic images-based deep learning model for detection with nasopharyngeal malignancies. Cancer Commun. 2018, 38, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, S.; Hou, J.; Yu, H.; Zhao, X.; Sun, Y.; Lambo, R.L.; Xie, Y.; Liu, L.; Qin, W.; Luo, W. Computer-Aided Pathologic Diagnosis of Nasopharyngeal Carcinoma Based on Deep Learning. Am. J. Pathol. 2020, 190, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Chuang, W.-Y.; Chang, S.-H.; Yu, W.-H.; Yang, C.-K.; Yeh, C.-J.; Ueng, S.-H.; Liu, Y.-J.; Chen, T.-D.; Chen, K.-H.; Hsieh, Y.-Y.; et al. Successful Identification of Nasopharyngeal Carcinoma in Nasopharyngeal Biopsies Using Deep Learning. Cancers 2020, 12, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, L.M.; King, A.D.; Ai, Q.Y.H.; Lam, W.K.J.; Poon, D.M.C.; Ma, B.B.Y.; Chan, K.C.A.; Mo, F.K.F. Convolutional neural network for discriminating Nasopharyngeal carcinoma and benign hyperplasia on MRI. Eur. Radiol. 2021, 31, 3856–3863. [Google Scholar] [CrossRef]
- Ke, L.; Deng, Y.; Xia, W.; Qiang, M.; Chen, X.; Liu, K.; Jing, B.; He, C.; Xie, C.; Guo, X.; et al. Development of a self-constrained 3D DenseNet model in automatic detection and segmentation of nasopharyngeal carcinoma using magnetic resonance images. Oral Oncol. 2020, 110, 104862. [Google Scholar] [CrossRef] [PubMed]
- Men, K.; Chen, X.; Zhang, Y.; Zhang, T.; Dai, J.; Yi, J.; Li, Y. Deep Deconvolutional Neural Network for Target Segmentation of Nasopharyngeal Cancer in Planning Computed Tomography Images. Front. Oncol. 2017, 7, 315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Xu, Y.; Chen, Z.; Liu, D.; Feng, S.-T.; Law, M.; Ye, Y.; Huang, B. Tumor Segmentation in Contrast-Enhanced Magnetic Resonance Imaging for Nasopharyngeal Carcinoma: Deep Learning with Convolutional Neural Network. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zu, C.; Hu, G.; Luo, Y.; Ma, Z.; He, K.; Wu, X.; Zhou, J. Automatic Tumor Segmentation with Deep Convolutional Neural Networks for Radiotherapy Applications. Neural Process. Lett. 2018, 48, 1323–1334. [Google Scholar] [CrossRef]
- Ma, Z.; Wu, X.; Sun, S.; Xia, C.; Yang, Z.; Li, S.; Zhou, J. A discriminative learning based approach for automated Nasopharyngeal carcinoma segmentation leveraging multi-modality similarity metric learning. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 813–816. [Google Scholar]
- Daoud, B.; Morooka, K.; Kurazume, R.; Leila, F.; Mnejja, W.; Daoud, J. 3D segmentation of nasopharyngeal carcinoma from CT images using cascade deep learning. Comput. Med. Imaging Graph. 2019, 77, 101644. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Dou, Q.; Jin, Y.-M.; Zhou, G.-Q.; Tang, Y.-Q.; Chen, W.-L.; Su, B.-A.; Liu, F.; Tao, C.-J.; Jiang, N.; et al. Deep Learning for Automated Contouring of Primary Tumor Volumes by MRI for Nasopharyngeal Carcinoma. Radiology 2019, 291, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Tang, F.; Huang, X.; Yang, K.; Zhong, T.; Hu, R.; Liu, S.; Yuan, X.; Zhang, Y. Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur. Radiol. 2018, 29, 1961–1967. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Huang, X.; Tang, F.; Liang, S.; Deng, X.; Zhang, Y. Boosting-based cascaded convolutional neural networks for the segmentation of CT organs-at-risk in Nasopharyngeal carcinoma. Med. Phys. 2019, 46, 5602–5611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Zhou, S.; Wu, X.; Zhang, H.; Yan, W.; Sun, S.; Zhou, J. Nasopharyngeal carcinoma segmentation based on enhanced convolutional neural networks using multi-modal metric learning. Phys. Med. Biol. 2018, 64, 025005. [Google Scholar] [CrossRef]
- Li, S.; Xiao, J.; He, L.; Peng, X.; Yuan, X. The Tumor Target Segmentation of Nasopharyngeal Cancer in CT Images Based on Deep Learning Methods. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Qin, N.; Hao, X.; Shi, J.; Wu, A.; An, H.; Zhang, H.; Wu, A.; Yang, Y. Sequential and Iterative Auto-Segmentation of High-Risk Clinical Target Volume for Radiotherapy of Nasopharyngeal Carcinoma in Planning CT Images. Front. Oncol. 2020, 10, 1134. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qi, Y.; Yin, Y.; Li, T.; Liu, X.; Li, X.; Gong, G.; Wang, L. MMFNet: A multi-modality MRI fusion network for segmentation of nasopharyngeal carcinoma. Neurocomputing 2020, 394, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Shi, C.; Li, X.; Wu, X.; Zhou, J.; Lv, J. Image segmentation of nasopharyngeal carcinoma using 3D CNN with long-range skip connection and multi-scale feature pyramid. Soft Comput. 2020, 24, 12671–12680. [Google Scholar] [CrossRef]
- Ye, Y.; Cai, Z.; Huang, B.; He, Y.; Zeng, P.; Zou, G.; Deng, W.; Chen, H.; Huang, B. Fully-Automated Segmentation of Nasopharyngeal Carcinoma on Dual-Sequence MRI Using Convolutional Neural Networks. Front. Oncol. 2020, 10, 166. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Peng, H.; Dan, T.; Hu, Y.; Tao, G.; Cai, H. Coarse-to-fine Nasopharyngeal carcinoma Segmentation in MRI via Multi-stage Rendering. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 623–628. [Google Scholar]
- Jin, Z.; Li, X.C.; Shen, L.; Lang, J.; Li, J.; Wu, J.; Xu, P.; Duan, J. Automatic Primary Gross Tumor Volume Segmentation for Nasopharyngeal carcinoma using ResSE-UNet. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Mayo Clinic, Rochester, MN, USA, 28–30 July 2020; pp. 585–590. [Google Scholar]
- Wang, X.; Yang, G.; Zhang, Y.; Zhu, L.; Xue, X.; Zhang, B.; Cai, C.; Jin, H.; Zheng, J.; Wu, J.; et al. Automated delineation of nasopharynx gross tumor volume for nasopharyngeal carcinoma by plain CT combining contrast-enhanced CT using deep learning. J. Radiat. Res. Appl. Sci. 2020, 13, 568–577. [Google Scholar] [CrossRef]
- Wong, L.M.; Ai, Q.Y.H.; Mo, F.K.F.; Poon, D.M.C.; King, A.D. Convolutional neural network in nasopharyngeal carcinoma: How good is automatic delineation for primary tumor on a non-contrast-enhanced fat-suppressed T2-weighted MRI? Jpn. J. Radiol. 2021, 39, 571–579. [Google Scholar] [CrossRef]
- Bai, X.; Hu, Y.; Gong, G.; Yin, Y.; Xia, Y. A deep learning approach to segmentation of nasopharyngeal carcinoma using computed tomography. Biomed. Signal. Process. Control. 2021, 64, 102246. [Google Scholar] [CrossRef]
- Shboul, Z.; Alam, M.; Vidyaratne, L.; Pei, L.; Elbakary, M.I.; Iftekharuddin, K.M. Feature-Guided Deep Radiomics for Glioblastoma Patient Survival Prediction. Front. Neurosci. 2019, 13, 966. [Google Scholar] [CrossRef]
- Paul, R.; Hawkins, S.H.; Schabath, M.B.; Gillies, R.J.; Hall, L.O.; Goldgof, D.B. Predicting malignant nodules by fusing deep features with classical radiomics features. J. Med. Imaging 2018, 5, 011021. [Google Scholar] [CrossRef]
- Bizzego, A.; Bussola, N.; Salvalai, D.; Chierici, M.; Maggio, V.; Jurman, G.; Furlanello, C. Integrating deep and radiomics features in cancer bioimaging. In Proceedings of the 2019 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Certosa di Pontignano, Siena–Tuscany, Italy, 9–11 July 2019; pp. 1–8. [Google Scholar]
- Hatt, M.; Parmar, C.; Qi, J.; El Naqa, I. Machine (Deep) Learning Methods for Image Processing and Radiomics. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 104–108. [Google Scholar] [CrossRef]
- Li, S.; Wang, K.; Hou, Z.; Yang, J.; Ren, W.; Gao, S.; Meng, F.; Wu, P.; Liu, B.; Liu, J.; et al. Use of Radiomics Combined with Machine Learning Method in the Recurrence Patterns After Intensity-Modulated Radiotherapy for Nasopharyngeal Carcinoma: A Preliminary Study. Front. Oncol. 2018, 8, 648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, L.-Z.; Fang, X.-L.; Dong, D.; Peng, H.; Fang, M.-J.; Huang, C.-L.; He, B.-X.; Lin, L.; Ma, J.; Tang, L.-L.; et al. A deep learning MR-based radiomic nomogram may predict survival for Nasopharyngeal carcinoma patients with stage T3N1M0. Radiother. Oncol. 2020, 151, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhong, L.-Z.; Zhao, X.; Dong, D.; Yao, J.-J.; Wang, S.-Y.; Liu, Y.; Zhu, D.; Wang, Y.; Wang, G.-J.; et al. A deep-learning-based prognostic nomogram integrating microscopic digital pathology and macroscopic magnetic resonance images in nasopharyngeal carcinoma: A multi-cohort study. Ther. Adv. Med. Oncol. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coroller, T.P.; Bi, W.L.; Huynh, E.; Abedalthagafi, M.; Aizer, A.A.; Greenwald, N.F.; Parmar, C.; Narayan, V.; Wu, W.W.; Miranda de Moura, S.; et al. Radiographic prediction of meningioma grade by semantic and radiomic features. PLoS ONE 2017, 12, e0187908. [Google Scholar] [CrossRef] [Green Version]
- Kebir, S.; Khurshid, Z.; Gaertner, F.C.; Essler, M.; Hattingen, E.; Fimmers, R.; Scheffler, B.; Herrlinger, U.; Bundschuh, R.A.; Glas, M. Unsupervised consensus cluster analysis of [18F]-fluoroethyl-L-tyrosine positron emission tomography identified textural features for the diagnosis of pseudoprogression in high-grade glioma. Oncotarget 2016, 8, 8294–8304. [Google Scholar] [CrossRef]
- Antropova, N.; Huynh, B.Q.; Giger, M.L. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med. Phys. 2017, 44, 5162–5171. [Google Scholar] [CrossRef]
- Gierach, G.L.; Li, H.; Loud, J.T.; Greene, M.H.; Chow, C.K.; Lan, L.; Prindiville, S.A.; Eng-Wong, J.; Soballe, P.W.; Giambartolomei, C.; et al. Relationships between computer-extracted mammographic texture pattern features and BRCA1/2 mutation status: A cross-sectional study. Breast Cancer Res. 2014, 16, 424. [Google Scholar]
- Ciompi, F.; Chung, K.; Van Riel, S.J.; Setio, A.A.A.; Gerke, P.K.; Jacobs, C.; Scholten, E.T.; Schaefer-Prokop, C.; Wille, M.M.W.; Marchianò, A.; et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci. Rep. 2017, 7, 46479. [Google Scholar] [CrossRef]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Velazquez, E.R.; Leijenaar, R.T.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef]
- Zeng, Y.; Xu, S.; Chapman, W.C.; Li, S.; Alipour, Z.; Abdelal, H.; Chatterjee, D.; Mutch, M.; Zhu, Q. Real-time colorectal cancer diagnosis using PR-OCT with deep learning. Theranostics 2020, 10, 2587–2596. [Google Scholar] [CrossRef]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef] [Green Version]
- Spadarella, G.; Calareso, G.; Garanzini, E.; Ugga, L.; Cuocolo, A.; Cuocolo, R. MRI based radiomics in nasopharyngeal cancer: Systematic review and perspectives using radiomic quality score (RQS) assessment. Eur. J. Radiol. 2021, 140, 109744. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Collins, F.S. Precision medicine in 2030—seven ways to transform healthcare. Cell 2021, 184, 1415–1419. [Google Scholar] [CrossRef]
- Fan, M.; Xia, P.; Clarke, R.; Wang, Y.; Li, L. Radiogenomic signatures reveal multiscale intratumour heterogeneity associated with biological functions and survival in breast cancer. Nat. Commun. 2020, 11, 4861. [Google Scholar] [CrossRef]
- Iwatate, Y.; Hoshino, I.; Yokota, H.; Ishige, F.; Itami, M.; Mori, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; et al. Radiogenomics for predicting p53 status, PD-L1 expression, and prognosis with machine learning in pancreatic cancer. Br. J. Cancer 2020, 123, 1253–1261. [Google Scholar] [CrossRef]
- Shui, L.; Ren, H.; Yang, X.; Li, J.; Chen, Z.; Yi, C.; Zhu, H.; Shui, P. Era of radiogenomics in precision medicine: An emerging approach for prediction of the diagnosis, treatment and prognosis of tumors. Front. Oncol. 2020, 10, 3195. [Google Scholar]
- Jain, R.; Chi, A.S. Radiogenomics identifying important biological pathways in gliomas. Neuro-Oncology 2021, 23, 177–178. [Google Scholar] [CrossRef] [PubMed]
- Cho, N. Breast Cancer Radiogenomics: Association of Enhancement Pattern at DCE MRI with Deregulation of mTOR Pathway. Radiology 2020, 296, 288–289. [Google Scholar] [CrossRef]
- Pinker-Domenig, K.; Chin, J.; Melsaether, A.N.; Morris, E.A.; Moy, L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology 2018, 287, 732–747. [Google Scholar] [CrossRef]
- Badic, B.; Tixier, F.; Le Rest, C.C.; Hatt, M.; Visvikis, D. Radiogenomics in Colorectal Cancer. Cancers 2021, 13, 973. [Google Scholar] [CrossRef]
- Zhou, M.; Leung, A.; Echegaray, S.; Gentles, A.; Shrager, J.B.; Jensen, K.C.; Berry, G.J.; Plevritis, S.K.; Rubin, D.L.; Napel, S.; et al. Non–Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology 2018, 286, 307–315. [Google Scholar] [CrossRef]
- Vargas, H.A.; Huang, E.P.; Lakhman, Y.; Ippolito, J.E.; Bhosale, P.; Mellnick, V.; Shinagare, A.B.; Anello, M.; Kirby, J.; Fevrier-Sullivan, B.; et al. Radiogenomics of high-grade serous ovarian cancer: Multireader multi-institutional study from the Cancer Genome Atlas Ovarian Cancer Imaging Research Group. Radiology 2017, 285, 482–492. [Google Scholar] [CrossRef] [Green Version]
- Panayides, A.S.; Pattichis, M.S.; Leandrou, S.; Pitris, C.; Constantinidou, A.; Pattichis, C.S. Radiogenomics for precision medicine with a big data analytics perspective. IEEE J. Biomed. Health Inform. 2018, 23, 2063–2079. [Google Scholar] [CrossRef] [PubMed]
- Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease; National Academies Press: Washington, DC, USA, 2011.
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging imaging and genomics. Abdom. Radiol. 2019, 44, 1960–1984. [Google Scholar] [CrossRef] [Green Version]
- Arimura, H.; Soufi, M.; Kamezawa, H.; Ninomiya, K.; Yamada, M. Radiomics with artificial intelligence for precision medicine in radiation therapy. J. Radiat. Res. 2019, 60, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Lao, J.; Chen, Y.; Li, Z.-C.; Li, Q.; Zhang, J.; Liu, J.; Zhai, G. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci. Rep. 2017, 7, 10353. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Li, C.-L.; Luo, X.-M.; Chuan, Z.-R.; Lv, W.-Z.; Li, X.; Cui, X.-W.; Dietrich, C.F. Ultrasound-based deep learning radiomics in the assessment of pathological complete response to neoadjuvant chemotherapy in locally advanced breast cancer. Eur. J. Cancer 2021, 147, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Gospodarowicz, M.K.; Miller, D.; Groome, P.A.; Greene, F.L.; Logan, P.A.; Sobin, L.H.; Project, F.T.U.T. The process for continuous improvement of the TNM classification. Cancer 2003, 100. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J.W.L. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [PubMed] [Green Version]

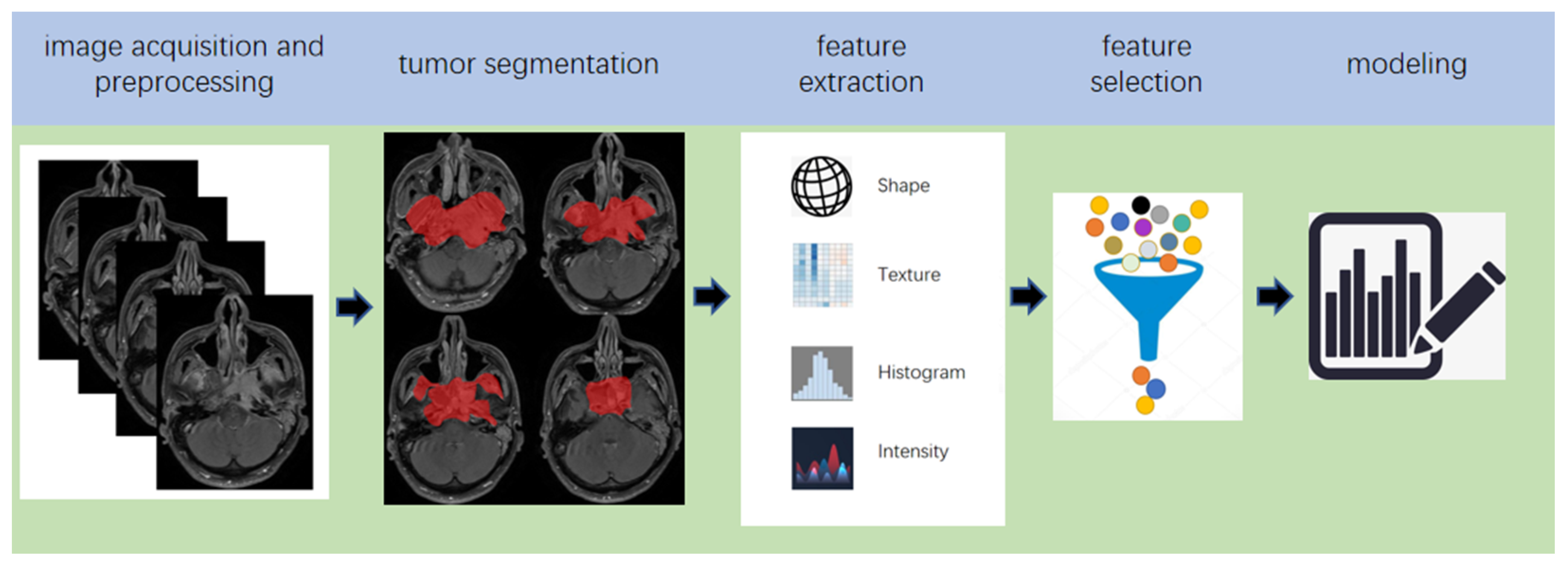




| Criterias | Detailed Rules and Regulations |
|---|---|
| Inclusion Criteria |
|
| Exclusion Criteria |
|
| Author, Year, Reference | Image | Sample Size (Patient) | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Zhang, B. (2017) [24] | MRI | 108 | LASSO | CR, nomograms, calibration curves | C-index 0.776 |
| Zhang, B. (2017) [81] | MRI | 110 | L1-LOG, L1-SVM, RF, DC, EN-LOG, SIS | L2-LOG, KSVM, AdaBoost, LSVM, RF, Nnet, KNN, LDA, NB | AUC 0.846 |
| Zhang, B. (2017) [82] | MRI | 113 | LASSO | RS | AUC 0.886 |
| Ouyang, F.S. (2017) [83] | MRI | 100 | LASSO | RS | HR 7.28 |
| Lv, W. (2019) [84] | PET/CT | 128 | Univariate analysis with FDR, SC > 0.8 | CR | C-index 0.77 |
| Zhuo, E.H. (2019) [85] | MRI | 658 | Entropy-based consensus clustering method | SVM | C-index 0.814 |
| Zhang, L.L. (2019) [86] | MRI | 737 | RFE | CR and nomogram | C-index 0.73 |
| Yang, K. (2019) [87] | MRI | 224 | LASSO | CR and nomogram | C-index 0.811 |
| Ming, X. (2019) [88] | MRI | 303 | Non-negative matrix factorization | Chi-squared test, nomogram | C-index 0.845 |
| Zhang, L. (2019) [89] | MRI | 140 | LR-RFE | CR and nomogram | C-index 0.74 |
| Mao, J. (2019) [90] | MRI | 79 | Univariate analyses | CR | AUC 0.825 |
| Du, R. (2019) [91] | MRI | 277 | Hierarchal clustering analysis, PR | SVM | AUC 0.8 |
| Xu, H. (2020) [92] | PET/CT | 128 | Univariate CR, PR > 0.8 | CR | C-index 0.69 |
| Shen, H. (2020) [93] | MRI | 327 | LASSO, RFE | CR, RS | C-index 0.874 |
| Bologna, M. (2020) [94] | MRI | 136 | Intra-class correlation coefficient, SCC > 0.85 | CR | C-index 0.72 |
| Feng, Q. (2020) [95] | PET/MR | 100 | LASSO | CR | AUC 0.85 |
| Peng, L. (2021) [96] | PET/CT | 85 | W-test, Chi-square test, PR, RA | SFFS coupled with SVM | AUC 0.829 |
| Author, Year, Reference | Image | Sample Size | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Zhang, L. (2019) [97] | MRI | 176 | LASSO | LR | AUC 0.792 |
| Zhong, X. (2020) [98] | MRI | 46 | LASSO | Nomogram | AUC 0.72 |
| Akram, F. (2020) [99] | MRI | 14 | Paired t-test and W-test | Shapiro-Wilk normality tests | p < 0.001 |
| Zhang, X. (2020) [100] | MRI | 238 | MRMR combined with 0.632 + bootstrap algorithms | RF | AUC 0.845 |
| Peng, L. (2021) [96] | PET/CT | 85 | W-test, PR, RA, Chi-square test | SFFS coupled with SVM | AUC 0.829 |
| Author, Year, Reference | Image | Sample Size | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Lv, W. (2018) [101] | PET/CT | 106 | Intra-class coefficient | LR with LOOCV | AUC 0.89 |
| Du, D. (2020) [102] | PET/CT | 76 | MIM, FSCR, RELF-F, MRMR, CMIM, JMI, SC > 0.7 | DT, KNN, LDA, LR, NB, RF, and SVM with radial basis function kernel | AUC 0.892 |
| Author, Year, Reference | Image | Sample Size | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Wang, G. (2018) [103] | MRI | 120 | LASSO | LR | AUC 0.822 |
| Yu, T.T. (2019) [104] | MRI | 70 | LASSO | Univariate LR | AUC 0.852 |
| Yongfeng, P. (2020) [105] | MRI | 108 | ANOVA/MW test, correlation analysis and LASSO | Multivariate LR | AUC 0.905 |
| Zhang, L. (2020) [106] | MRI | 265 | LASSO | LR | AUC 0.886, 0.863 |
| Zhao, L. (2020) [107] | MRI | 123 | t-test and LASSO based on LOOCV | SVM, nomogram, backward stepwise LR | C-index 0.863 |
| Author, Year, Reference | Image | Sample Size | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Liu, Y. (2019) [108] | CT | 35 | RFE | LR | Precision 0.922 |
| Zhang, B. (2020) [109] | MRI | 242 | The relief algorithm | RF | AUC 0.830 |
| Author, Year, Reference | Image | Sample Size | Modeling | Model Evaluation |
|---|---|---|---|---|
| Qiang, M.Y. (2019) [110] | MRI | 1636 | 3D DenseNet | HR 0.62 |
| Du, R. (2019) [111] | MRI | 596 | DCNN | AUC 0.69 |
| Yang, Q. (2020) [112] | MRI | 1138 | Resnet network | AUC 0.943 |
| Jing, B. (2020) [113] | MRI | 1417 | Multi-modality deep survival network | C-index 0.651 |
| Qiang, M. (2020) [34] | MRI | 3444 | 3D-CNN | C-index 0.776 |
| Cui, C. (2020) [114] | MRI | 792 | Automatic machine learning (AutoML) including DL | AUC 0.796 |
| Liu, K. (2020) [115] | Pathology | 1055 | Neural network DeepSurv | C-index 0.723 |
| Zhang, L. (2021) [116] | MRI | 233 | Resnet network | AUC 0.808 |
| Author, Year, Reference | Image | Sample Size | Modeling | Model Evaluation |
|---|---|---|---|---|
| Li, Y. (2019) [117] | CBCT | 70 | U-Net neural network (DCNN) | 1%/1 mm GPR 95.5% |
| Wang, Y. (2019) [118] | MRI | 33 | U-Net neural network (DCNN) | MAE: 97 ± 13 HU in soft tissue, 131 ± 24 HU in all region, 357 ± 44 HU in bone |
| Tie, X. (2020) [119] | MRI | 32 | ResU-Net | Structural similarity index 0.92 |
| Peng, Y. (2020) [120] | MRI | 173 | GANs | 2%/2mm GPR 98.52~98.68% |
| Author, Year, Reference | Image | Sample Size | Modeling | Model Evaluation |
|---|---|---|---|---|
| Mohammed, M.A. (2018) [121,122,123] | Endoscopic images | 381 images | ANN | Accuracy 96.22% |
| Li, C. (2018) [124] | Endoscopic images | 28,966 images | Fully convolutional network (FCNN) | Accuracy 88.7% |
| Diao, S. (2020) [125] | Pathology | 731 patients | Inception-v3 | AUC 0.936 |
| Chuang, W.Y. (2020) [126] | Pathology | 726 patients | ResNeXt | AUC 0.985 |
| Wong, L.M. (2020) [127] | MRI | 412 patients | Residual Attention Network (RAN) | AUC 0.96 |
| Ke, L. (2020) [128] | MRI | 4100 patients | 3D DenseNet | Accuracy 97.77% |
| Author, Year, Reference | Image | Sample Size | Modeling | Model Evaluation |
|---|---|---|---|---|
| Men, K. (2017) [129] | CT | 230 | DDNN and VGG-16 | DSC GTVnx 80.9% GTVnd 62.3% CTV 82.6% |
| Li, Q. (2018) [130] | MRI | 29 | CNN | DSC 0.89 |
| Wang, Y. (2018) [131] | MRI | 15 | DCNN | DSC 0.79 |
| Ma, Z. (2018) [132] | CT, MRI | 50 | Multi-modality CNN | DSC 0.636 (CT), 0.712 (MRI) |
| Daoud, B. (2019) [133] | CT | 70 | CNN | DSC 0.91 |
| Lin, L. (2019) [134] | MRI | 1021 | 3D-CNN | DSC 0.79 |
| Liang, S. (2019) [135] | CT | 185 | CNN | DSC 0.689–0.937 |
| Zhong, T. (2019) [136] | CT | 140 | CNN | DSC Parotids 0.92 Thyroids 0.92 Optic nerves 0.89 |
| Ma, Z. (2019) [137] | CT, MRI | 90 | Single-modality CNN multi-modality CNN | DSC 0.746 (CT), 0.752 (MRI) |
| Li, S. (2019) [138] | CT | 502 | U-Net (CNN) | DSC Lymph nodes 0.659 Tumor 0.74 |
| Xue, X. (2020) [139] | CT | 150 | SI-Net and U-Net | DSC 0.84 |
| Chen, H. (2020) [140] | MRI | 149 | 3D-CNN | DSC 0.724 |
| Guo, F. (2020) [141] | MRI | 120 | 3D-CNN | DSC 0.737 |
| Ye, Y. (2020) [142] | MRI | 44 | Dense connectivity embedding U-net | DSC 0.87 |
| Li, Y. (2020) [143] | MRI | 596 | ResNet-101 | DSC 0.703 |
| Jin, Z. (2020) [144] | CT | 90 | ResSE-UNet | DSC 0.84 |
| Wang, X. (2020) [145] | CT | 205 | 3D U-Net | DSC 0.827 |
| Ke, L. (2020) [128] | MRI | 4100 | 3D DenseNet | DSC 0.77 |
| Wong, L.M. (2021) [146] | MRI | 195 | CNN | DSC 0.73 |
| Bai, X. (2021) [147] | MRI | 60 | ResNeXt-50 and U-Net | DSC 0.618 |
| Author, Year, Reference | Image | Sample Size | Feature Selection | Modeling | Model Evaluation |
|---|---|---|---|---|---|
| Li, S. (2018) [152] | CT | 306 | ICC, PCC, and PCA | ANN, KNN, SVM | AUC ANN: 0.812 KNN: 0.775 SVM: 0.732 |
| Peng, H. (2019) [30] | PET/CT | 707 | LASSO | DCNNs and nomogram | C-index 0.722 |
| Zhong, L.Z. (2020) [153] | MRI | 638 | Not described | SE-ResNeXt, CR and nomogram | C-index 0.788 |
| Zhang, F. (2020) [154] | MRI, Pathology | 220 | ICC > 0.75, Univariate analysis, MRMR, RF | ResNet-18, Nomogram | C-index 0.834 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Deng, Y.-Q.; Zhu, Z.-L.; Hua, H.-L.; Tao, Z.-Z. A Comprehensive Review on Radiomics and Deep Learning for Nasopharyngeal Carcinoma Imaging. Diagnostics 2021, 11, 1523. https://doi.org/10.3390/diagnostics11091523
Li S, Deng Y-Q, Zhu Z-L, Hua H-L, Tao Z-Z. A Comprehensive Review on Radiomics and Deep Learning for Nasopharyngeal Carcinoma Imaging. Diagnostics. 2021; 11(9):1523. https://doi.org/10.3390/diagnostics11091523
Chicago/Turabian StyleLi, Song, Yu-Qin Deng, Zhi-Ling Zhu, Hong-Li Hua, and Ze-Zhang Tao. 2021. "A Comprehensive Review on Radiomics and Deep Learning for Nasopharyngeal Carcinoma Imaging" Diagnostics 11, no. 9: 1523. https://doi.org/10.3390/diagnostics11091523






