State-of-the-Art Techniques for Diagnosis of Medical Parasites and Arthropods
Abstract
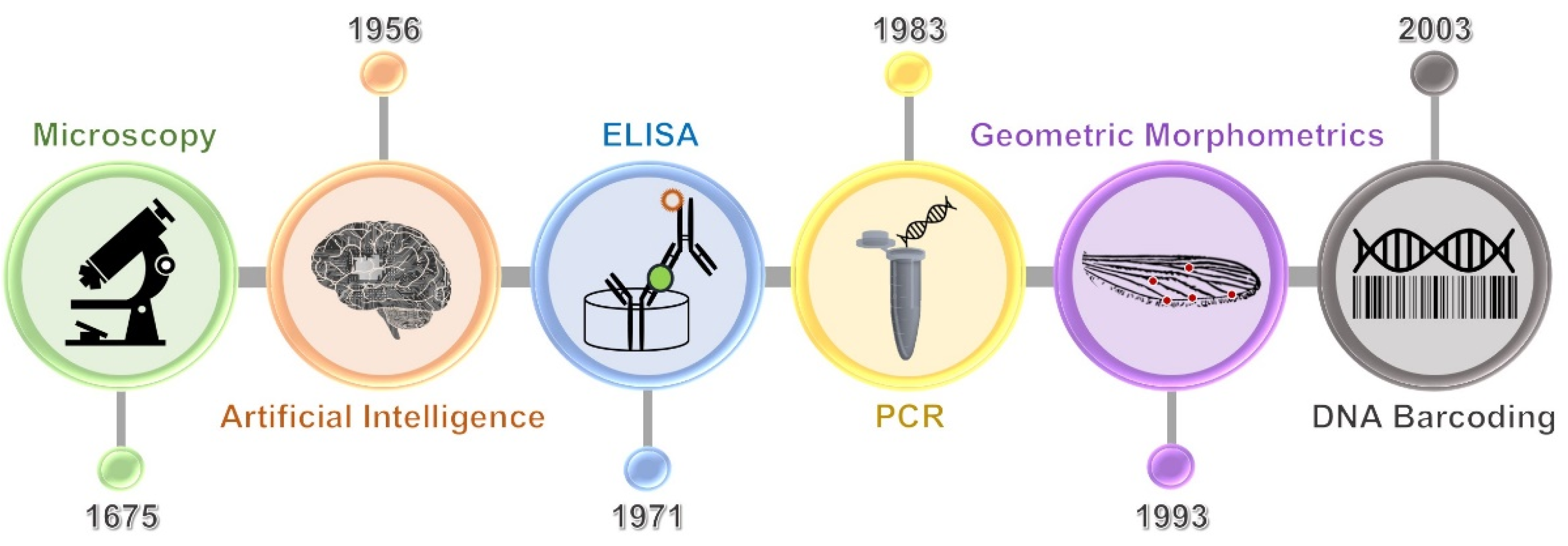
:1. Introduction
2. Conventional Diagnostic Methods for Medical Parasite and Arthropod
2.1. Gross Examination
2.2. Microscopic Examination
| Types of Examination | Method Names | Diagnostic Tool of | References | |
|---|---|---|---|---|
| Parasite 1 | Arthropod 2 | |||
| Gross | Gross examination | Yes | Yes | [10,11] |
| Microscopic | Direct wet smear | Yes | No | [14] |
| Concentration technique | Yes | No | [15] | |
| Staining technique | Yes | No | [16] | |
| Scotch tape technique | Yes | No | [17] | |
| Skin scraping technique | No | Yes | [18] | |
| Culture | Harada–Mori technique | Yes | No | [21] |
| Baermann technique | Yes | No | [22] | |
| Charcoal culture technique | Yes | No | [23] | |
| Agar plate culture technique | Yes | No | [24] | |
| Immunological | Enzyme-linked immunosorbent assay (ELISA) | Yes | No | [25] |
| Immunoblot assay | Yes | No | [25] | |
| Molecular biology | Polymerase chain reaction (PCR) | Yes | Yes | [12,26,27] |
| Loop-mediated isothermal amplification (LAMP) | Yes | Yes | [20,26,28] | |
| xMAP assay (Luminex) | Yes | No | [26,29] | |
| Restriction fragment length polymorphism (RFLP) | Yes | Yes | [26,30] | |
| Next-generation sequencing (NGS) | Yes | Yes | [20,31] | |
| Mass spectrometry (MS) | Yes | Yes | [12,32] | |
| Biosensors | Yes | No | [20,33] | |
2.3. Culture Technique
2.4. Immunological Examination
2.5. Molecular Biology Examination
3. Advanced Approaches for Medical Parasite and Arthropod Diagnoses
3.1. DNA Barcoding Technique
3.2. Geometric Morphometric Analysis
3.3. Artificial Intelligence Technology
4. Future Perspectives and Challenges of Parasite and Arthropod Diagnostics
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tungtrongchitr, A. Host-parasite relationship. In Medical Helminthology, 2nd ed.; Bhaibulaya, M., Ed.; Wattanakijpanich Publishers: Bangkok, Thailand, 2017; pp. 6–11. [Google Scholar]
- Singh, B.; Varikuti, S.; Halsey, G.; Volpedo, G.; Hamza, O.M.; Satoskar, A.R. Host-directed therapies for parasitic diseases. Future Med. Chem. 2019, 11, 1999–2018. [Google Scholar] [CrossRef]
- Taghipour, A.; Olfatifar, M.; Rostami, A.; Foroutan, M.; Vasigala, V.; Norouzi, M. Intestinal parasites in hemodialysis patients from developing countries: A systematic review and meta-analysis. Hemodial. Int. 2020, 24, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wongsaroj, T.; Nithikathkul, C.; Rojkitikul, W.; Nakai, W.; Royal, L.; Rammasut, P. National survey of helminthiasis in Thailand. Asian Biomed. 2014, 8, 779–783. [Google Scholar] [CrossRef] [Green Version]
- Chonsawat, P.; Wongphan, B. Prevalence of parasitic infections in patients at Hospital for Tropical Diseases, Mahidol University. J. Med. Tech. Assoc. Thail. 2017, 45, 6073–6084. [Google Scholar]
- Eldridge, B.F.; Edman, J.D. Introduction to medical entomology. In Medical Entomology: A Textbook on Public Health and Veterinary Problems Caused by Arthropods, 2nd ed.; Eldridge, B.F., Edman, J.D., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 1–12. [Google Scholar]
- Raza, N.; Qadir, S.N.; Agha, H. Risk factors for scabies among male soldiers in Pakistan: Case-control study. East. Mediterr. Health J. 2009, 15, 1105–1110. [Google Scholar] [CrossRef]
- Hatam-Nahavandi, K.; Ahmadpour, E.; Pashazadeh, F.; Dezhkam, A.; Zarean, M.; Rafiei-Sefiddashti, R.; Salimi-Khorashad, A.; Hosseini-Teshnizi, S.; Hazratian, T.; Otranto, D. Pediculosis capitis among school-age students worldwide as an emerging public health concern: A systematic review and meta-analysis of past five decades. Parasitol. Res. 2020, 119, 3125–3143. [Google Scholar] [CrossRef]
- Haq, K.A.U.; Gul, N.A.; Hammad, H.M.; Bibi, Y.; Bibi, A.; Mohsan, J. Prevalence of Giardia intestinalis and Hymenolepis nana in Afghan refugee population of Mianwali district, Pakistan. Afr. Health Sci. 2015, 15, 394–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrivastav, S.; Jha, A. Paragonimiasis: A missed diagnosis from Nepal. Respir. Med. Case Rep. 2020, 31, 101298. [Google Scholar] [PubMed]
- Baker, D.J.; Kantor, G.R.; Stierstorfer, M.B.; Brady, G. Furuncular myiasis from Dermatobia hominis infestation. Diagnosis by light microscopy. Am. J. Dermatopathol. 1995, 17, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Yansouni, C.P.; Merckx, J.; Libman, M.D.; Ndao, M. Recent advances in clinical parasitology diagnostics. Curr. Infect. Dis. Rep. 2014, 16, 434. [Google Scholar] [CrossRef]
- Garcia, L.S. Diagnostic Medical Parasitology, 4th ed.; ASM Press: Washington, DC, USA, 2001; pp. 741–785. [Google Scholar]
- Neimeister, R.; Logan, A.L.; Egleton, J.H.; Kleger, B. Evaluation of direct wet mount parasitological examination of preserved fecal specimens. J. Clin. Microbiol. 1990, 28, 1082–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, J.A.; Estevez, E.G. Method for concentration of parasites from small amounts of feces. J. Clin. Microbiol. 1983, 18, 786–788. [Google Scholar] [CrossRef] [Green Version]
- Gomori, G. A rapid one-step trichrome stain. Am. J. Clin. Pathol. 1950, 20, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.F. A device for the diagnosis of Enterobius infection. Am. J. Trop. Med. 1941, 21, 159–161. [Google Scholar] [CrossRef]
- Zorbozan, O.; Türk, B.G.; Acar, A.; Oraloğlu, G.; Ünver, A.; Töz, S.; Ünal, İ.; Turgay, N. Comparison of skin scraping and standard superficial skin biopsy in the laboratory diagnosis of scabies. Turk. Parazitol. Derg. 2020, 44, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.E.C.; Cardoso, A.E.O.; Talhari, C.; Santos, M. Update on parasitic dermatoses. An. Bras. Dermatol. 2020, 95, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Paparini, A.; Oskam, C. New technologies for detection of enteric parasites. Trends Parasitol. 2017, 33, 532–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harada, U.; Mori, O. A new method for culturing hookworm. Yonago Acta Med. 1955, 1, 177–179. [Google Scholar]
- Baermann, G. A Simple Method for the Detection of Ankylostomum (Nematode) Larvae in Soil Tests; Javasche Boekhandel & Drukkerij: Batavia, Germany, 1917; p. 41. [Google Scholar]
- Shorb, D.A. A method of separating infective larvae of Haemonchus contortus (Trichostrongylidae) from free living nematodes. In Proceedings of the Helminthological Society of Washington, Washington, DC, USA, 6 August 1937; Volume 4, p. 52. [Google Scholar]
- Arakaki, T.; Iwanaga, M.; Kinjo, F.; Saito, A.; Asato, R.; Ikeshiro, T. Efficacy of agar-plate culture in detection of Strongyloides stercoralis infection. J. Parasitol. 1990, 76, 425–428. [Google Scholar] [CrossRef]
- Ndao, M. Diagnosis of parasitic diseases: Old and new approaches. Interdiscip. Perspect. Infect. Dis. 2009, 2009, 278246. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.G.; Staggemeier, R.; Borges, A.L.P.; Rodrigues, M.T.; Castelan, L.A.; Vasconcelos, J.; Anschau, M.E.; Spalding, S.M. Molecular techniques for the study and diagnosis of parasite infection. J. Venom. Anim. Toxins Incl. Trop. Dis. 2011, 17, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Bae, M.; Kim, J.Y.; Jung, J.; Cha, H.H.; Jeon, N.Y.; Lee, H.J.; Kim, M.J.; Chang, S.E.; Kim, S.H. Diagnostic value of the molecular detection of Sarcoptes scabiei from a skin scraping in patients with suspected scabies. PLoS Negl. Trop. Dis. 2020, 14, e0008229. [Google Scholar] [CrossRef]
- Huang, B.; Montgomery, B.L.; Adamczyk, R.; Ehlers, G.; van den Hurk, A.F.; Warrilow, D. A LAMP-based colorimetric assay to expedite field surveillance of the invasive mosquito species Aedes aegypti and Aedes albopictus. PLoS Negl. Trop. Dis. 2020, 14, e0008130. [Google Scholar] [CrossRef]
- Reslova, N.; Michna, V.; Kasny, M.; Mikel, P.; Kralik, P. xMAP technology: Applications in detection of pathogens. Front. Microbiol. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Oduwole, O.A.; Oringanje, C.M.; Oduola, A.O.; Nwachuku, N.S.; Meremikwu, M.M.; Alaribe, A.A.A. Species composition of Anopheles (Diptera: Culicidae) in selected forested tourist areas of Nigeria endemic for malaria. J. Med. Entomol. 2020, 57, 2007–2010. [Google Scholar] [CrossRef]
- Zhang, X.M.; Shi, Z.Y.; Zhang, S.Q.; Zhang, P.; Wilson, J.J.; Shih, C.; Li, J.; Li, X.D.; Yu, G.Y.; Zhang, A.B. Plant-herbivorous insect networks: Who is eating what revealed by long barcodes using high-throughput sequencing and Trinity assembly. Insect Sci. 2021, 28, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, J.; Roesler, U. MALDI-TOF MS profiling-advances in species identification of pests, parasites, and vectors. Front. Cell Infect. Microbiol. 2017, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Luka, G.; Samiei, E.; Dehghani, S.; Johnson, T.; Najjaran, H.; Hoorfar, M. Label-free capacitive biosensor for detection of Cryptosporidium. Sensors 2019, 19, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, N.H. Cultivation of parasites. Trop. Parasitol. 2014, 4, 80–89. [Google Scholar] [CrossRef]
- Wheeler, R.J.; Gluenz, E.; Gull, K. The limits on Trypanosomatid morphological diversity. PLoS ONE 2013, 8, e79581. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Hua, Y.; Hu, G.L.; Hua, B.Z. Habitat divergence shapes the morphological diversity of larval insects: Insights from scorpionflies. Sci. Rep. 2019, 9, 12708. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; De Waard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Ondrejicka, D.A.; Locke, S.A.; Morey, K.; Borisenko, A.V.; Hanner, R.H. Status and prospects of DNA barcoding in medically important parasites and vectors. Trends Parasitol. 2014, 30, 582–591. [Google Scholar] [CrossRef]
- Morand, S. Advances and challenges in barcoding of microbes, parasites, and their vectors and reservoirs. Parasitology 2018, 145, 537–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Gjerde, B. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of Toxoplasma gondii, Neospora caninum, Hammondia heydorni and Hammondia triffittae (Apicomplexa: Sarcocystidae). Parasitol. Res. 2013, 112, 1493–1511. [Google Scholar]
- Fet, V.; Graham, M.R.; Webber, M.M.; Blagoev, G. Two new species of Euscorpius (Scorpiones: Euscorpiidae) from Bulgaria, Serbia, and Greece. Zootaxa 2014, 3894, 83–105. [Google Scholar] [CrossRef] [Green Version]
- Ondrejicka, D.A.; Morey, K.C.; Hanner, R.H. DNA barcodes identify medically important tick species in Canada. Genome 2017, 60, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Yang, Y.; Zhao, Y.; Niu, D.; Yang, R.; Wang, R.; Lu, Z.; Li, X. DNA barcoding for molecular identification of Demodex based on mitochondrial genes. Parasitol. Res. 2017, 116, 3285–3290. [Google Scholar] [CrossRef]
- Sumruayphol, S.; Siribat, P.; Dujardin, J.P.; Dujardin, S.; Komalamisra, C.; Thaenkham, U. Fasciola gigantica, F. hepatica and Fasciola intermediate forms: Geometric morphometrics and an artificial neural network to help morphological identification. PeerJ 2020, 8, e8597. [Google Scholar] [CrossRef] [Green Version]
- Hugot, J.P.; Baylac, M. Shape patterns of genital papillae in pinworms (Enterobiinae, Oxyurida, Nematoda) parasite of primates: A landmark analysis. Infect. Genet. Evol. 2007, 7, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Mondal, R.; Devi, N.P.; Jauhari, R.K. Landmark-based geometric morphometric analysis of wing shape among certain species of Aedes mosquitoes in District Dehradun (Uttarakhand), India. J. Vector Borne Dis. 2015, 52, 122–128. [Google Scholar] [PubMed]
- Sungvornyothin, S.; Kumlert, R.; Paris, D.H.; Prasartvit, A.; Sonthayanon, P.; Apiwathnasorn, C.; Morand, S.; Stekolnikov, A.A.; Sumruayphol, S. Geometric morphometrics of the scutum for differentiation of trombiculid mites within the genus Walchia (Acariformes: Prostigmata: Trombiculidae), a probable vector of scrub typhus. Ticks Tick Borne Dis. 2019, 10, 495–503. [Google Scholar] [CrossRef]
- Dujardin, J.P.; Le Pont, F. Morphometrics of a neotropical sandfly subspecies, Lutzomyia carrerai thula. C. R. Acad. Sci. III 2000, 323, 273–279. [Google Scholar] [CrossRef]
- Henriques, D.; Chávez-Galarza, J.; Teixeira, J.S.G.; Ferreira, H.; Neves, C.J.; Francoy, T.M.; Pinto, M.A. Wing geometric morphometrics of workers and drones and single nucleotide polymorphisms provide similar genetic structure in the Iberian honey bee (Apis mellifera iberiensis). Insects 2020, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Friedman, N.R.; Lecroq Bennet, B.; Fischer, G.; Sarnat, E.M.; Huang, J.P.; Knowles, L.L.K.; Economo, E.P. Macroevolutionary integration of phenotypes within and across ant worker castes. Ecol. Evol. 2020, 10, 9371–9383. [Google Scholar] [CrossRef] [PubMed]
- Josek, T.; Allan, B.F.; Alleyne, M. Morphometric analysis of chemoreception organ in male and female ticks (Acari: Ixodidae). J. Med. Entomol. 2018, 55, 547–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santillán-Guayasamín, S.; Villacís, A.G.; Grijalva, M.J.; Dujardin, J.P. The modern morphometric approach to identify eggs of Triatominae. Parasites Vectors 2017, 10, 55. [Google Scholar] [CrossRef] [Green Version]
- Poostchi, M.; Silamut, K.; Maude, R.J.; Jaeger, S.; Thoma, G. Image analysis and machine learning for detecting malaria. Transl. Res. 2018, 194, 36–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, K.; Bachman, C.M.; Delahunt, C.B.; Baldeon, J.A.; Alava, F.; Vilela, D.G.; Proux, S.; Mehanian, C.; McGuire, S.K.; Thompson, C.M.; et al. Automated microscopy for routine malaria diagnosis: A field comparison on Giemsa-stained blood films in Peru. Malar. J. 2018, 17, 339. [Google Scholar] [CrossRef] [Green Version]
- Holmström, O.; Linder, N.; Ngasala, B.; Mårtensson, A.; Linder, E.; Lundin, M.; Moilanen, H.; Suutala, A.; Diwan, V.; Lundin, J. Point-of-care mobile digital microscopy and deep learning for the detection of soil-transmitted helminths and Schistosoma haematobium. Glob. Health Action 2017, 10, 1337325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathison, B.A.; Kohan, J.L.; Walker, J.F.; Smith, R.B.; Ardon, O.; Couturier, M.R. Detection of intestinal protozoa in trichrome-stained stool specimens by use of a deep convolutional neural network. J. Clin. Microbiol. 2020, 58, e02053-19. [Google Scholar] [CrossRef] [Green Version]
- Kittichai, V.; Pengsakul, T.; Chumchuen, K.; Samung, Y.; Sriwichai, P.; Phatthamolrat, N.; Tongloy, T.; Jaksukam, K.; Chuwongin, S.; Boonsang, S. Deep learning approaches for challenging species and gender identification of mosquito vectors. Sci. Rep. 2021, 11, 4838. [Google Scholar] [CrossRef] [PubMed]
- Nesi, N.; Nakouné, E.; Cruaud, C.; Hassanin, A. DNA barcoding of African fruit bats (Mammalia, Pteropodidae). The mitochondrial genome does not provide a reliable discrimination between Epomophorus gambianus and Micropteropus pusillus. C. R. Biol. 2011, 334, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F.J.; Marcus, L.F. A revolution morphometrics. Trends Ecol. Evol. 1993, 8, 129–132. [Google Scholar] [CrossRef]
- Adams, D.C.; Rohlf, F.J.; Slice, D.E. Geometric morphometrics: Ten years of progress following the ‘revolution’. Ital. J. Zool. (Modena) 2004, 71, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Dujardin, S.; Dujardin, J.P. Geometric morphometrics in the cloud. Infect. Genet. Evol. 2019, 70, 189–196. [Google Scholar] [CrossRef]
- XYOM-CLIC: Morphometrics in Medical Entomology—Collection of Landmark for Identification and Characterization. Available online: https://xyom-clic.eu/the-clic-package (accessed on 14 August 2021).
- García-Sánchez, A.M.; Reguera-Gomez, M.; Valero, M.A.; Cutillas, C. Differentiation of Trichuris species eggs from non-human primates by geometric morphometric analysis. Int. J. Parasitol. Parasites Wildl. 2020, 12, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, J.P. Morphometrics applied to medical entomology. Infect. Genet. Evol. 2008, 8, 875–890. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69S, S36–S40. [Google Scholar] [CrossRef]
- Smith, K.P.; Kirby, J.E. Image analysis and artificial intelligence in infectious disease diagnostics. Clin. Microbiol. Infect. 2020, 26, 1318–1323. [Google Scholar] [CrossRef]
- Egli, A.; Schrenzel, J.; Greub, G. Digital microbiology. Clin. Microbiol. Infect. 2020, 26, 1324–1331. [Google Scholar] [CrossRef]
- Smith, K.P.; Kang, A.D.; Kirby, J.E. Automated interpretation of blood culture Gram stains by use of a deep convolutional neural network. J. Clin. Microbiol. 2018, 56, e01521-17. [Google Scholar] [CrossRef] [Green Version]
- Croxatto, A.; Marcelpoil, R.; Orny, C.; Morel, D.; Prod’hom, G.; Greub, G. Towards automated detection, semi-quantification and identification of microbial growth in clinical bacteriology: A proof of concept. Biomed. J. 2017, 40, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, A.; Acar, S.; Körkoca, H. Characterization of Klebsiella isolates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and determination of antimicrobial resistance with VITEK 2 advanced expert system (AES). Turk. J. Med. Sci. 2015, 45, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, J.T.; Ouattara, M.; D’Ambrosio, M.V.; Fletcher, D.A.; Keiser, J.; Utzinger, J.; N’Goran, E.K.; Andrews, J.R.; Bogoch, I.I. Accuracy of mobile phone and handheld light microscopy for the diagnosis of schistosomiasis and intestinal protozoa infections in Côte d’Ivoire. PLoS Negl. Trop. Dis. 2016, 10, e0004768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stemple, C.C.; Angus, S.V.; Park, T.S.; Yoon, J.Y. Smartphone-based optofluidic lab-on-a-chip for detecting pathogens from blood. J. Lab. Autom. 2014, 19, 35–41. [Google Scholar] [CrossRef]
- Franssen, F.; Janse, I.; Janssen, D.; Caccio, S.M.; Vatta, P.; van der Giessen, J.; van Passel, M. Mining public metagenomes for environmental surveillance of parasites: A proof of principle. Front. Microbiol. 2021, 12, 622356. [Google Scholar] [CrossRef]
- Bassene, H.; Niang, E.H.A.; Fenollar, F.; Dipankar, B.; Doucouré, S.; Ali, E.; Michelle, C.; Raoult, D.; Sokhna, C.; Mediannikov, O. 16S Metagenomic comparison of Plasmodium falciparum-infected and noninfected Anopheles gambiae and Anopheles funestus microbiota from Senegal. Am. J. Trop Med. Hyg. 2018, 99, 1489–1498. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Weng, X.; Xu, C.; Lin, Y.; Cheng, C.; Wei, H.; Chen, W. Metagenomic next-generation sequencing as a diagnostic tool for toxoplasmic encephalitis. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Giordano, F.; Ning, Z. Oxford nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.C.; Islam, M.M.; Jack Li, Y.C. Development of user-friendly tools for biomedical research and healthcare. Comput. Methods Programs Biomed. 2018, 167, A1. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of care testing for infectious diseases. Clin. Chim. Acta 2019, 493, 138–147. [Google Scholar] [CrossRef] [PubMed]



| Methods | Principle | References |
|---|---|---|
| DNA barcoding technique | Analysis of barcode sequence | [38,39,43,44,45] |
| Geometric morphometric analysis | Statistical analysis of shape pattern variation of an anatomical structure | [46,47,48,49,50,51,52,53,54] |
| Artificial intelligence technology | Analysis of picture using the trained algorithms | [55,56,57,58,59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruenchit, P. State-of-the-Art Techniques for Diagnosis of Medical Parasites and Arthropods. Diagnostics 2021, 11, 1545. https://doi.org/10.3390/diagnostics11091545
Ruenchit P. State-of-the-Art Techniques for Diagnosis of Medical Parasites and Arthropods. Diagnostics. 2021; 11(9):1545. https://doi.org/10.3390/diagnostics11091545
Chicago/Turabian StyleRuenchit, Pichet. 2021. "State-of-the-Art Techniques for Diagnosis of Medical Parasites and Arthropods" Diagnostics 11, no. 9: 1545. https://doi.org/10.3390/diagnostics11091545






