Classification of Sleep Apnea Based on Sub-Band Decomposition of EEG Signals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Database Description
2.1.1. ISRUC Sleep Database
2.1.2. Sleep—EDF Database
2.1.3. CAP Sleep Database
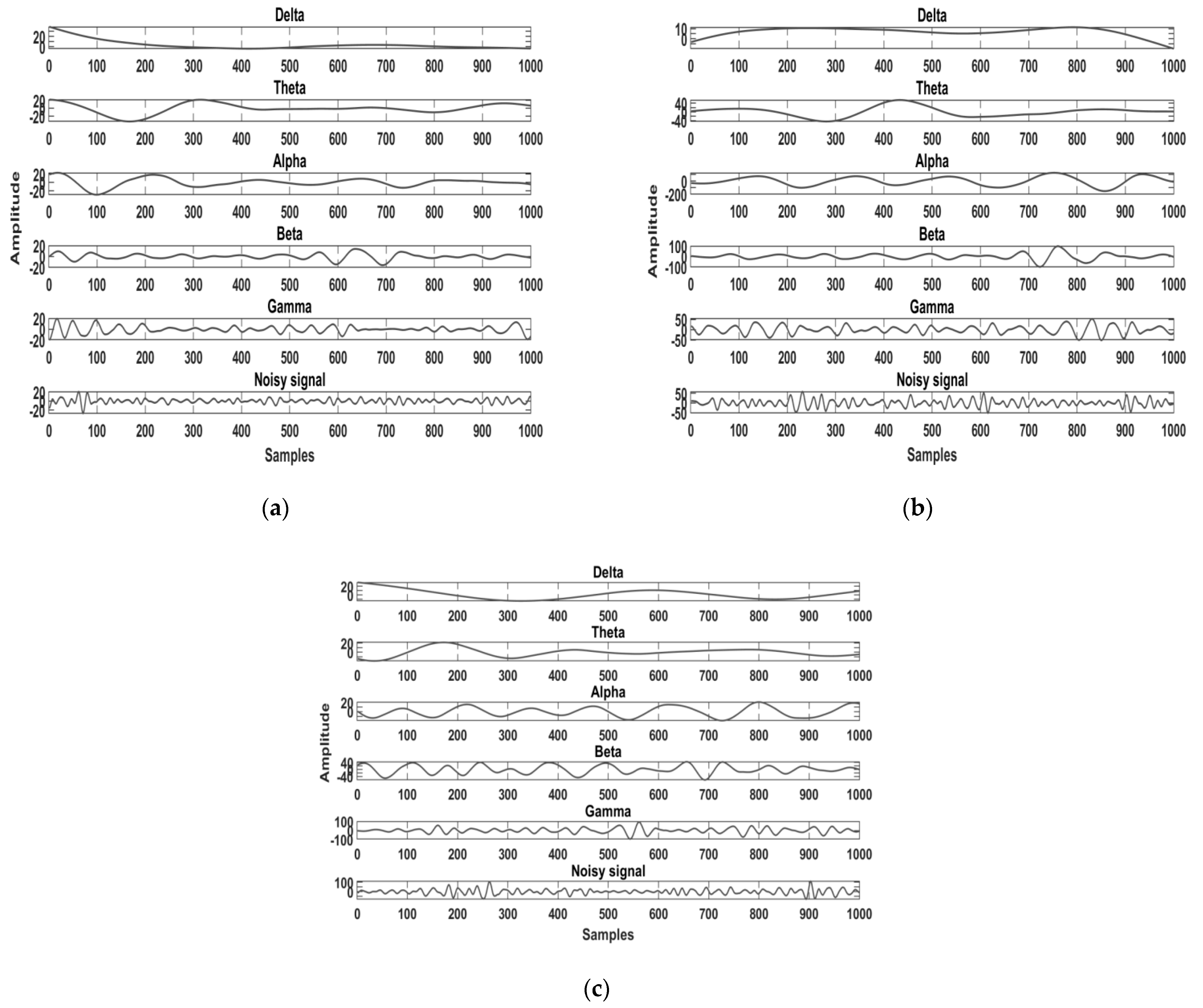
2.2. Pre-Processing and Band Separation
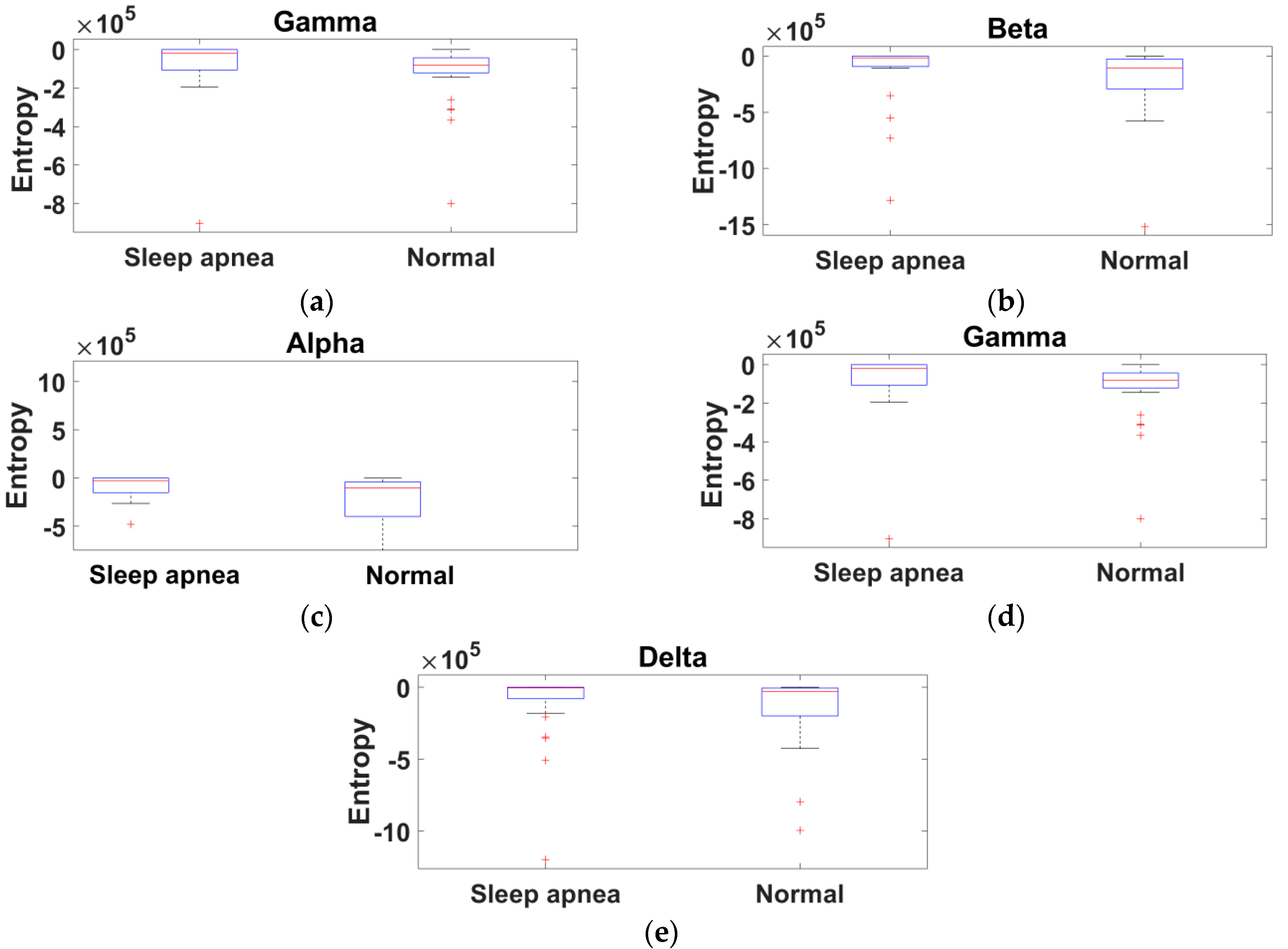
2.3. Feature Extraction Method of the Proposed Study
2.4. Classification Module
2.4.1. Support Vector Machine Classifier with Kernels
2.4.2. Random Forest Classifier
2.5. Performance Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, B.P. Indian Insurance Sector Must Step in to Cover the Cost of Sleep Studies. Available online: https://health.economictimes.indiatimes.com/news/industry/74200624 (accessed on 19 February 2020).
- Castro, I.D.; Varon, C.; Torfs, T.; Van Huffel, S.; Puers, R.; Van Hoof, C. Evaluation of a multichannel non-contact ECG system and signal quality algorithms for sleep apnea detection and monitoring. Sensors 2018, 18, 577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goyal, M.; Johnson, J. Obstructive sleep apnea diagnosis and management. Mo. Med. 2017, 114, 120–124. [Google Scholar]
- Tan, H.-L.; Gozal, D.; Ramirez, H.M.; Bandla, H.P.R.; Kheirandish-Gozal, L. Overnight polysomnography versus respiratory polygraphy in the diagnosis of pediatric obstructive sleep apnea. Sleep 2014, 37, 255–260. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Agarwal, S.; Acharya, U.R. Application of an optimal class of antisymmetric wavelet filter banks for obstructive sleep apnea diagnosis using ECG signals. Comput. Biol. Med. 2018, 100, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, Y.; Wang, J. A RR interval based automated apnea detection approach using residual network. Comput. Methods Programs Biomed. 2019, 176, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Zheng, X.; Shao, X.; Cui, L. Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity 2018, 2018, 9248410. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Li, Y.; Wen, P.P. Analysis and classification of sleep stages based on difference visibility graphs from a single channel EEG Signal. IEEE J. Biomed. Health Inform. 2014, 18, 1813–1821. [Google Scholar] [CrossRef]
- Sharma, M.; Goyal, D.; Achuth, P.; Acharya, U.R. An accurate sleep stages classification system using a new class of optimally time-frequency localized three-band wavelet filter bank. Comput. Biol. Med. 2018, 98, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Tzimourta, D.; Tsilimbaris, A.; Tzioukalia, K.; Tzallas, A.T.; Tsipouras, M.G.; Astrakas, L.G.; Giannakeas, N. EEG-Based automatic sleep stage classification. Biomed. J. Sci. Tech. Res. 2018, 7, 6032–6037. [Google Scholar]
- Ghasemzadeh, P.; Kalbkhani, H.; Shayesteh, M.G. Sleep stages classification from EEG signal based on Stockwell transform. IET Signal Process. 2019, 13, 242–252. [Google Scholar] [CrossRef]
- Savareh, B.A.; Bashiri, A.; Behmanesh, A.; Meftahi, G.H.; Hatef, B. Performance comparison of machine learning techniques in sleep scoring based on wavelet features and neighboring component analysis. PeerJ 2018, 6, e5247. [Google Scholar]
- Boostani, R.; Karimzadeh, F.; Nami, M. A comparative review on sleep stage classification methods in patients and healthy individuals. Comput. Methods Programs Biomed. 2017, 140, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Hertenstein, E.; Gabryelska, A.; Spiegelhalder, K.; Nissen, C.; FJohann, A.; Umarova, R.; Riemann, D.; Baglioni, C.; Fiege, F. Reference data for polysomnography-measured and subjective sleep in healthy adults. J. Clin. Sleep Med. 2018, 14, 523–534. [Google Scholar] [CrossRef]
- Gabryelska, A.; Feige, B.; Riemann, D.; Spiegehalder, K.; Johann, A.; Białasiewicz, P.; Hertenstein, E. Can spectral power predict subjective sleep quality in healthy individuals? J. Sleep Res. 2019, 28, e12848. [Google Scholar] [CrossRef] [PubMed]
- Elwali, A.; Moussavi, Z. A Novel decision making procedure during wakefulness for screening obstructive sleep apnea using anthropometric information and tracheal breathing sounds. Sci. Rep. 2019, 9, 11467. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, G.; Kanagasabai, A.; Mohan, J.; Seshadri, N.G. Music induced emotion using wavelet packet decomposition—An EEG study. Biomed. Signal Process. Control 2018, 42, 115–128. [Google Scholar] [CrossRef]
- Almuhammadi, W.S.; Aboalayon, K.A.I.; Faezipour, M. Efficient obstructive sleep apnea classification based on EEG signals, 2015 Long Island Systems. Appl. Technol. 2015, 15295284. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, X.; Yang, T.; Ji, S.; Wang, H.; Wang, J.; Wang, Y.; Yu, Q. Classification of sleep apnea based on EEG sub-band signal characteristics. Sci. Rep. 2021, 11, 5811–5824. [Google Scholar]
- Saha, S.; Bhattacharjee, A.; Fattah, S.A. Automatic detection of sleep apnea events based on inter-band energy ratio obtained from multi-band EEG signal. Healthc. Technol. Lett. 2019, 6, 82–86. [Google Scholar] [CrossRef]
- Nagendra, H.; Vinod Kumar Mukherjee, S. Cognitive behavior evaluation based on the physiological parameters among young healthy subjects with yoga as intervention. Comput. Math. Methods Med. 2015, 2015, 821061. [Google Scholar] [CrossRef]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef]
- Hassan, A.R.; Bhuiyan, M.I.H. Automated identification of sleep states from EEG signals by means of ensemble empirical mode decomposition and random under sampling boosting. Comput. Methods Programs Biomed. 2017, 140, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Dhok, S.; Pimpalkhute, V.; Chandurkar, A.; Bhurane, A.A.; Sharma, M.; Acharya, U.R. Automated phase classification in cyclic alternating patterns in sleep stages using Wigner–Ville Distribution based features. Comput. Biol. Med. 2020, 119, 103691. [Google Scholar] [CrossRef] [PubMed]
- Meddins, R. Introduction to Digital Signal Processing; Newnes: Oxford, UK, 2000. [Google Scholar]
- Ravikumar, K.; Tamilselvan, S. On the use of wavelets packet decomposition for time series prediction. Appl. Math. Sci. 2014, 8, 2847–2858. [Google Scholar] [CrossRef]
- Kufoy, E.; Palma, J.-A.; Lopez, J.; Alegre, M.; Urrestarazu, E.; Artieda, J.; Iriarte, J. Changes in the heart rate variability in patients with obstructive sleep apnea and its response to acute CPAP treatment. PLoS ONE 2013, 7, e33769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswabhargav, C.S.; Tripathy, R.K.; Acharya, U.R. Automated detection of sleep apnea using sparse residual entropy features with various dictionaries extracted from heart rate and EDR signals. Comput. Biol. Med. 2019, 108, 20–30. [Google Scholar] [CrossRef]
- Marcal, T.A.; dos Santos, J.M.; Rosa, A.; Cardoso, J.M. OSAS assessment with entropy analysis of high resolution snoring audio signals. Biomed. Signal Process. Control 2020, 61, 101965. [Google Scholar] [CrossRef]
- Mohanavelu, K.; Poonguzhali, S.; Adalarasu, K.; Ravi, D.; Chinnadurai, V.; Vinutha, S.; Ramachandran, K.; Jayaraman, S. Dynamic cognitive workload assessment of fighter pilots in simulated fighter aircraft environment using EEG. Biomed. Signal Process. Control 2020, 61, 102018. [Google Scholar]
- Janbakhshi, P.; Shamsollahi, M. Sleep apnea detection from single-Lead ECG using features based on ECG-derived respiration (EDR) Signals. IRBM 2018, 39, 206–218. [Google Scholar] [CrossRef]
- Tripathy, R.; Gajbhiye, P.; Acharya, U.R. Automated sleep apnea detection from cardio-pulmonary signal using bivariate fast and adaptive EMD coupled with cross time–frequency analysis. Comput. Biol. Med. 2020, 120, 103769. [Google Scholar] [CrossRef]
- Sugianela, Y.; Sutino, Q.L.; Herumurti, D. EEG classification for epilepsy based on wavelet packet decomposition and random forest. J. Sci. Inf. 2018, 11, 27–33. [Google Scholar] [CrossRef]
- Rajeswari, J.; Jagannath, M. Screening of obstructive sleep apnea in an urban population in south India. Obes. Med. 2020, 18, 100220. [Google Scholar] [CrossRef]
- Ting, W.; Guo-Zheng, Y.; Bang-Hua, Y.; Hong, S. EEG feature extraction based on wavelet packet decomposition for brain computer interface. Measurement 2008, 41, 618–625. [Google Scholar] [CrossRef]
- Rachim, V.P.; Li, G.; Chung, W.-Y. Sleep apnea classification using ECG-signal wavelet-PCA features. Bio.-Med. Mater. Eng. 2014, 24, 2875–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.Q.; Hossen, A. Identification of obstructive sleep apnea using artificial neural networks and wavelet packet decomposition of the HRV signal. J. Eng. Res. 2020, 17, 24–33. [Google Scholar]
- Freeman, F.G.; Mikulka, P.J.; Prinzel, L.J.; Scerbo, M.W. Evaluation of an adaptive automation system using three EEG indices with a visual tracking task. Biol. Psychol. 1999, 50, 61–76. [Google Scholar] [CrossRef]
- Al-Angari, H.M.; Sahakian, A.V. Automated recognition of obstructive sleep apnea syndrome using support vector machine classifier. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 463–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| ISRUC | Sleep-EDF | Cap Sleep | ||||
|---|---|---|---|---|---|---|
| Subjects (M:F) | Age (Years) | Subjects (M:F) | Age (Years) | Subjects (M:F) | Age (Years) | |
| Sleep subjects | 57(33:24) | 55 ± 14 | 20(15:5) | 39 ± 18.54 | 4(4:0) | 71.25 ± 7.22 |
| Normal subjects | 32(18:14) | 45 ± 17 | 20(16:4) | 33.15 ± 9.37 | 16(7:9) | 32.18 ± 5.55 |
| Training set | 79 | - | 30 | - | 16 | - |
| Testing set | 10 | - | 10 | - | 4 | - |
| Authors | Signals Used | Feature Used | Database | Subjects Used | Classifier Used | Event | Accuracy |
|---|---|---|---|---|---|---|---|
| Castro et al. [2] | ECG | Heart rate Heart rate variability | Volunteers | 15 | Signal quality indication | Sleep apnea | 91.0% |
| Shrama et al. [5] | ECG | Fuzzy entropy, Log energy | Apnea ECG | 27 | Least square SVM | Sleep apnea | 90.0% |
| Wang et al. [6] | ECG | RR intervals | Apnea ECG | 35 | Residual network | Sleep apnea | 94.0% |
| Cui et al. [7] | EEG (F3-A2, C3-A2, O1-A2, F4-A1, C4-A1, and O2-A1) | Entropy | ISRUC | 116 | Convolutional neural network | Sleep stage classification (Wake, stage N1, stage N2, stage N3, and stage REM) | 92.2% |
| Zhu et al. [8] | EEG (Pz-Oz) | Degree distribution, Horizontal visual graph, Difference visual graph | Sleep-EDF | 8 | Support vector machine | Sleep stage classification | 87.5% |
| Tzimourta et al. [10] | EEG (F3-A2, C3-A2, O1-A2, F4-A1, C4-A1, and O2-A1) | Energy | ISRUC | 100 | Random forest | Sleep stage classification (Wake, stage N1, stage N2, stage N3, and stage REM) | 75.3% |
| Savareh et al. [12] | EEG (Fpz-Cz and Pz-Oz) | Wavelet tree features | Sleep-EDF | 61 | Support vector machine, Artificial neural network | Sleep stage classification | 90.3% ANN |
| Boostani et al. [13] | ECG | Entropy | Sleep-EDF | 20 | Random forest | Sleep apnea | 87.1% |
| Elwali and Moussavi [16] | ECG | Optimized set of breathing sounds | PSG Sleep database at Misericordia Health Center (Winnipeg, Canada) | 199 | Random forest | Sleep apnea | 81.4% |
| Aluhummadi et al. [18] | EEG | Energy, Variance | MIT-BIH | 18 | Support vector machine, Linear discriminant analysis, Naive Baiyes, Artificial neural network | Sleep apnea | 97% SVM |
| Zhao et al. [19] | EEG (C3-A2 and C4-A1) | Sample entropy Variance | Tianjin Chest Hospital | 30 | Support vector machine, K nearest neighbor, Random forest | Sleep apnea | 88.99% SVM |
| Saha et al. [20] | EEG (C3-A2 and C4-A1) | Inter band energy ratio δ-θ δ-α δ-σ δ-β θ-α | St. Vincent’s University Hospital/University College Dublin sleep apnea database | 5 | K nearest neighbor, Support vector machine, Linear discriminant analysis Naïve Bayes, | Sleep apnea | 91.6% KNN |
| Tripathy et al. [32] | ECG | Heart rate Respiration signals | Apnea ECG | 31 | Support vector machine, Random forest | Sleep apnea | 77.8% SVM |
| Rachim et al. [36] | ECG | Heart rate Respiration signals ECG-derived respiration | Apnea ECG | 35 | Support vector machine | Sleep apnea | 95.0% |
| Ali et al. [37] | ECG | Heart rate variability | Sultan Qaboos University Hospital (SQUH) | 80 | Support vector machine | Obstructive sleep apnea | 95.0% |
| Al-Angari et al. [39] | ECG | Respiration rate Oxygen saturation | Sleep Heart Health Study | 100 | Support vector machine | Obstructive sleep apnea | 95.0% |
| Proposed study | EEG (C3-A2, Fpz-Cz, Pz-Oz, and C4-A1) | Entropy Energy Heart rate Synchronization Neural activity Brain perfusion | ISRUC, Sleep-EDF, CAP Sleep | 159 | Support vector machine, Random forest | Sleep apnea | 90.0% SVM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayaraj, R.; Mohan, J. Classification of Sleep Apnea Based on Sub-Band Decomposition of EEG Signals. Diagnostics 2021, 11, 1571. https://doi.org/10.3390/diagnostics11091571
Jayaraj R, Mohan J. Classification of Sleep Apnea Based on Sub-Band Decomposition of EEG Signals. Diagnostics. 2021; 11(9):1571. https://doi.org/10.3390/diagnostics11091571
Chicago/Turabian StyleJayaraj, Rajeswari, and Jagannath Mohan. 2021. "Classification of Sleep Apnea Based on Sub-Band Decomposition of EEG Signals" Diagnostics 11, no. 9: 1571. https://doi.org/10.3390/diagnostics11091571
APA StyleJayaraj, R., & Mohan, J. (2021). Classification of Sleep Apnea Based on Sub-Band Decomposition of EEG Signals. Diagnostics, 11(9), 1571. https://doi.org/10.3390/diagnostics11091571






