A comprehensive ML pipeline is proposed in this study to select morphological features and brain regions that relates to ASD. The ML pipeline starts with downloading the sMRI volumes of ASD and TD subjects provided by ABIDE I dataset [
53], then the preprocessing of the sMRI volumes is performed by Freesurfer V.6.0 [
54,
55,
56,
57]. Preprocessing consists of three stages, which are: (i) intensity normalization, (ii) skull stripping, and (iii) brain segmentation. Each of the aforementioned stages comprises a set of substages, which are going to be briefly discussed in the following sections. After preprocessing, features are extracted in the form of two numerical representation for each morphological feature for each brain region. A data matrix, and a target vector are created and passed to feature selection algorithm to select the candidate imaging markers. Reduced data matrix based on the candidate imaging markers, and the target vector are then passed to the ML algorithms to select the best ML model that can be used for classifying ASD and TD subjects.
2.2. Pre-Processing
Preprocessing is a crucial requirement to eliminate the between-subjects variability that may stem from data acquisition, different scanners, artifacts, or partial volume effects. Moreover, brain MRI scans usually contain non-brain tissues as it is shown in
Figure 1. FreeSurfer performs multiple steps on each sMRI volume to extract the morphological features. Those steps are intensity normalization, brain extraction and skull stripping, brain segmentation and area labeling, tessellation of the gray-white matter boundary, surface inflation and spherical atlas registration, and eventually cortical surface parcellation to the Desikan–Killiany (DK) atlas.
It is worth noting that the main assumption behind the preprocessing is that as long as FreeSurfer succeeds in extracting the morphological features of the cerebral cortex and parcelate them to DK, then confounding variables relevant to the MRI scanner wont be major concern. This assumption is based on the fact that FreeSurfer outputs the morphological features in their physical unit e.g., mm, mm2, mm3.
2.2.1. Intensity Normalization
Variations in both intensity and contrast across sMRI images, resulting in the corruption of the sMRI images, are typically due to magnetic susceptibility artifacts and RF-field inhomogeneities. This corruption is undesirable for any segmentation procedure, which utilizes intensity information in order to classify voxel data into different tissue types [
63]. To correct the aforementioned corruption, the following procedure of 11 steps is repeated with iterating oversteps from (viii)–(x).
The procedures are: (i) Construct a set of histograms from overlapping slices parallel to the x-y Cartesian plane in the magnetic co-ordinate system. (ii) Smooth the resulting histograms using a fairly broad Gaussian window. (iii) Use a peak-finding algorithm to determine the mean white matter intensity. (iv) Discard the outliers from the array of the detected mean white matter intensities. (v) Fit a set of cubic splines to the resulting coefficients of the valid slices. (vi) Use the splines to interpolate the coefficients for each point along the
z axis. (vii) Adjust each intensity value by the coefficient at its z coordinate. (viii) Find all points in the volume that are at the center of a
neighborhood of intensity values that all lie within 10% of the white matter peak. (ix) Build a Voronoi diagram and set all voxels unassigned in step (viii) to the correction value of the nearest control point. (x) Perform a few iterations of “soap-bubble” smoothing. (xi) Scale the intensity at each voxel in the volume by the computed correction field [
64].
The results are shown in
Figure 1, the visual transformation of the brightness level between the preprocessing step and the normalization step is to reduce the variance of brightness for the same tissue inter-subjects due to different data acquisition methods. For more mathematical and implementation details, the reader is referred to [
64,
65].
2.2.2. Brain Extraction or Skull Stripping
Brain extraction of skull stripping is the process of automatically strip the skull (or any non-brain tissue) from the intensity normalized image. In order to remove the skull and any non-brain tissue, a tessellated ellipsoidal template is deformed into the shape of the inner surface in the skull. Two kind of forces drive the deformation process: (i) An MRI-based force, and (ii) A curvature reducing force.
The MRI-based force is designed to drive the template outward from the brain. It is calculated based on nonlocal information obtained by sampling the MRI data along the surface normal to each vertex of the template tessellation. The curvature reducing force enforces a smoothness constraint on the deformed template, which can be seen as encoding
a priori knowledge about the smoothness of the inner surface of the skull [
64]. The result of this step is illustrated in
Figure 1.
2.2.3. Brain Segmentation & Area Labeling
The segmentation process is a two-step procedure: (i) A preliminary classification is performed based solely on the intensity information, and (ii) This volume is examined and the regions that contain more than one tissue type are marked for further processing [
64]. After segmentation, a 3D surface reconstruction and brain parcellation to an anatomical atlas is performed on the segmented volume. The 3D surface reconstruction is performed via 2 steps: (i) tessellation of the gray-white matter boundary as described in [
54,
66], and (ii) surface inflation and spherical atlas registration as described in [
54,
65].
Brain parcellation to an anatomical atlas, which is the Desikan–Killiany (DK) atlas, is described in [
57]. DK atlas parcellates the brain into 68 cortical labels, 34 for each hemisphere. The results of the segmentation and the DK atlas parcellation are shown in
Figure 1. For more detailed information on each of the aforementioned preprocessing steps, the reader is referred to the following publication [
53].
2.3. Feature Extraction
There are two outputs of FreeSurfer, which are (i) a set of volumes for each subject describing each step of the pipeline (normalization, skull stripping and segmentation) as shown in
Figure 1, and (ii) surfaces parcellated to DK atlas and containing the morphological features values at each point on a predefined mesh grid created on the brain, as shown in
Figure 2.
In this study, we utilized the following morphological features to represent the brain of each subject: (i) surface area (
), (ii) volume (
V), (iii) thickness (
), and (iv) curvature (
c) (see
Figure 2). It is worth noting that
is calculated as the closest distance from the gray/white matter boundary to the gray/CSF boundary at each vertex on the tessellated surface [
67], while
c is measured as the average of the reciprocal of the principal radii [
57].
For each of those features, we calculated the median value (
), inter-quartile range (
), and
within each brain region parcellated to the DK atlas. There are two reasons behind choosing
and
to represent each morphological feature of each brain region: (i) the distribution of morphological features’ values within each brain region is not necessary Gaussian as it is shown in
Figure 3, and (ii) to include lower and upper bound that each morphological feature can possess within a specific brain region while excluding the outliers.
DK atlas parcellates the brain into 68 brain regions; 34 brain regions on the left hemisphere, and 34 brain regions on the right hemisphere. Therefore, each subject is represented by a vector of 68 brain regions × 4 features × 2 = 544 elements of a feature vector.
ABIDE I comprises 17 different sites with total number of 1112 subjects after performing quality control, and removing all subjects with bad brain segmentation, at which the data were collected. Thus, the data are heterogeneous, and it is invalid to assume blocking for all confounding variables while working on the whole dataset. Therefore, we proposed two exclusion criteria: (i) exclusion criterion for subjects, and (ii) exclusion criterion for sites.
The exclusion criterion for subjects is simply done by removing the subjects with missing feature values. The exclusion criterion for sites depends on how balanced each site is. In other words, after applying subjects’ excluding criterion, if we find a site where the ratio ASD:TD, or its reciprocal TD:ASD, exceeds 0.6, we discard that site.There is a trade-off between removing all the subjects of the unbalanced sites, or raise the unbalanced threshold. Empirically, we found that 0.6 would be a reasonable ratio, given that we utilize balanced accuracy score to evaluate the system performance, to assume balance and include as many sites as possible in the study.
The rationale behind the site’s exclusion criterion is to avoid including too many subjects of one class that have been collected with certain criterion without having their corresponding subjects from the other class that possess the same collection criterion, i.e., trying to avoid introducing more heterogeneity due to the subject’s exclusion criterion.
Table 1 shows the summary statistics of the data set after applying both exclusion criteria, subject’s exclusion criteria and site exclusion criteria. Total number of five sites have been discarded, which are: KKI, SDSU, NYU, SBL, and USM, representing a total of 305 subjects. Over the whole data set, there is no statistically significant difference between ASD and TD group
.
Furthermore, there is no statistically significant difference between the age of each group
. However, there is a statistically significant difference between the gender within each group; for TD group, chi-square test was conducted over the gender distribution
, and for the ASD group, the chi-square test was conducted over the gender distribution (
,
. At the end of this step, a data matrix is created as follows:
where
D is the data matrix with size 664 subjects
features; each row represents the feature vector of a specific subject.
denoted the feature value
j of subject
i, and
denoted the diagnosis of subject
i. It is worth mentioning that
D is the data matrix for the global model.
For the local model, we created 12 data matrices (
) such that
each corresponding to one of the sites;
denotes the data matrix corresponding to site
t. Each
has the size of
such that
M denotes the number of subjects within site t; sequentially,
denotes the diagnosis vector corresponding to site
t, and
denotes the set containing all the
for all sites.
2.4. Feature Adjustment & Normalization
As it has been mentioned in the literature that there is an effect of age on ASD brain morphology [
68], morphological features have been adjusted, for the effect of both age and sex, in the proposed work. Adjusted metrics of regional
V and
were calculated using cortical growth curves from Coupé et al. [
69]. Denote by
the mean volume of cortical grey matter in individuals of sex
s and age
a. Then each regional volume
is replaced by its age-relative, adjusted metric
. Similarly, each regional surface area
is converted to an adjusted metric
[
69].
The feature vector corresponding to every subject contains the
and
of each morphological feature for every region. We consider
and
to be the lower bound, and the upper bound of every morphological feature for every brain region respectively. Morphological features don’t share the same units of measurement; for instance, surface area is measured in
, while
V is measured in
. Consequently, we anticipate having different ranges of values, which might adversely affect the performance of the classifiers [
70].
In this study, we utilized minimum–maximum normalization between 0 to 1 as it is one of the most common normalization methods used for biomedical data [
71]. Consequently, each column in the data matrix
D is normalized between 0 to 1 using the Equation (
1).
where
and
denote the normalized feature value
j and the original feature value
j corresponding to the subject
i, and
and
correspond to minimum and maximum values of the feature vector
j respectively. The output normalized matrix is denoted by
for the global model, and
for the local model where
.
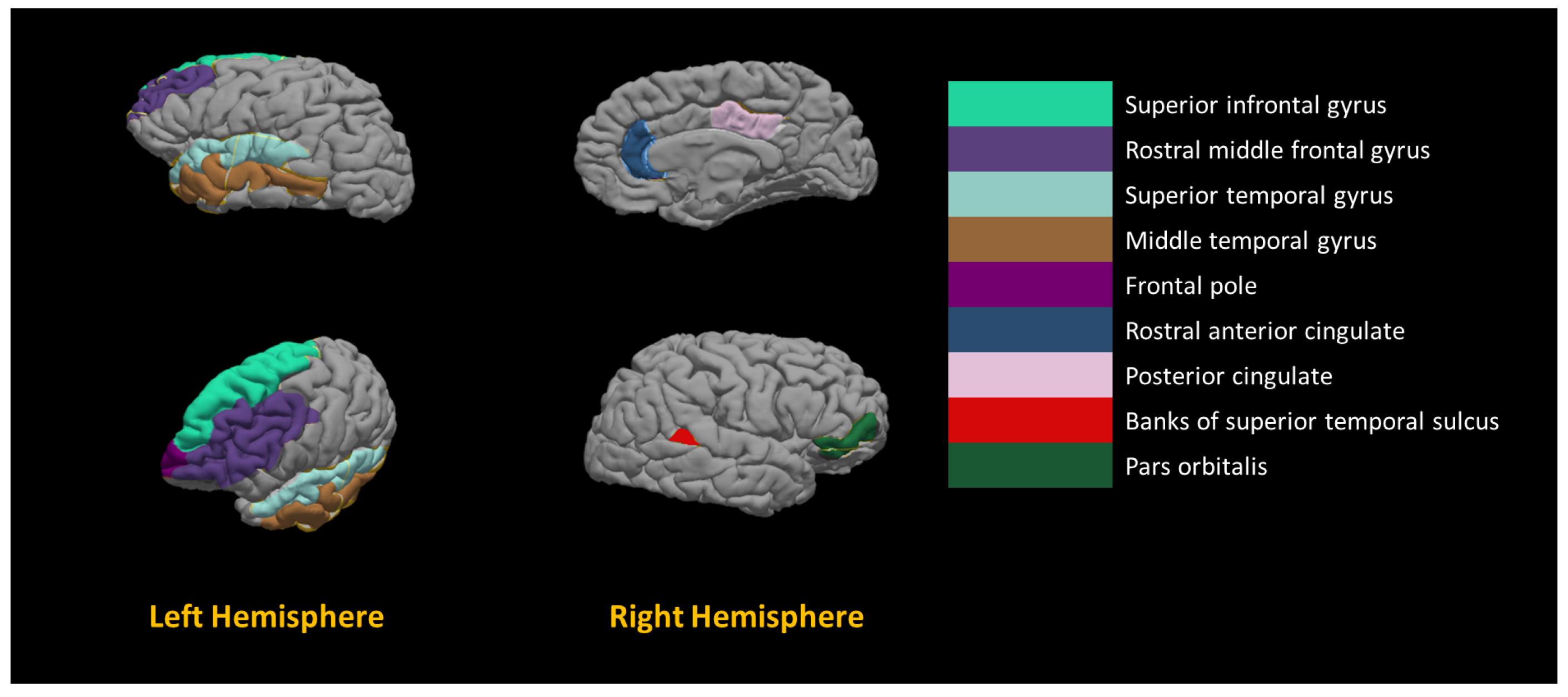
2.5. Building Neuro-Atlas
To implement a Computer-Aided Diagnosis System (CAD) for accurate diagnosis of autism, we have to use a neuro-atlas tailored to the specific developmental patterns of the brain in autism. Unfortunately, there is no general purpose brain atlas in the literature that we can use in our CAD system; thus, developing an atlas for autistic subjects that shows the areas and imaging markers that are associated with autism is the main motivation behind this work. To achieve this goal, we used the modern tools of machine learning (e.g., Recursive Feature Elimination via Cross Validation (RFECV)) to select the most significant features and their corresponding areas that are correlated with autism spectrum disorder.
Since RFECV is one of supervised feature selection algorithm, we have to split the data into
k-folds,
in our case (as shown on
Figure 4, and for each fold (i) train a predetermined classifier using the training set, (ii) evaluate the performance of the trained classifier on the validation set, (iii) save the classifier’s score on the validation set, (iv) find the least significant feature according to the trained classifier, (v) remove the least significant feature from the model, and (vi) repeat the whole process until you end up with only one feature.
Again, repeat the whole process for each fold, calculate the average performance of the cross-validation (CV) when: using all features to train the classifier; using all features but one, and so forth, to the point of classification on a single feature. Find the number of the features at which the classifier has the maximum performance score, assuming it is features. is the optimum number of features to be selected.
Perform the whole algorithm again over all the subjects to find the most
significant features. The algorithm is discussed in detail in Algorithm 1. For further details regarding the algorithm and its implementation, the reader is suggested to read Guyon et al. [
72] and Pedregosa et al. [
73], respectively.
To build a neuro-atlas for autism, ABIDE I dataset and RFECV are utilized to select those significant brain regions along with their morphological features. For both the local model and the global model, RFECV is run with four different classifier architectures, which are RF, least absolute shrinkage and selection operator (LASSO), RIDGE regression (RIDGE), and SVM with linear kernel, resulting in four different models.
Those four models represent two major categories of features’ sets: (i) A feature set that forms a feature space, where the subjects are non-linearly separable as much as possible, and (ii) A feature set that forms a feature space, where the subjects are linearly separable as much as possible. The first category corresponds to the features’ set selected by RFECV+RF, and the second category corresponds to the features’ sets selected by RFECV+LASSO, RFECV+RIDGE, RFECV+SVM.
Each of the RFECV models is performed with 10-fold CV; such that we iterate over all the 544 features, removing one feature at a time, perform 10-fold CV on the current sample, and calculate the average balanced accuracy score. The balanced accuracy score was introduced in 2010 to solve the optimistic estimate occurs when a biased classifier is tested on an imbalanced dataset [
74]. The balanced accuracy score is defined by Equation (
2):
where
denotes the balanced accuracy score,
denotes the true positive classified by the model,
denotes the total number of positive cases in the sample,
denotes the true negatives classified by the model, and
is the total number of negative cases in the sample.
| Algorithm 1: RFECV |
![Diagnostics 12 00165 i001 Diagnostics 12 00165 i001]() |
For the site-based model, RFECV is performed on each site separately. The selected set of features is then extracted for each site to have a new data matrix with number of columns less than or equal to the original number of columns. For the global model, RFECV is performed only one time on the normalized data matrix , and the selected set of features is calculated and then extracted from . At this point, we assume that the selected features from each site are the imaging markers candidate for ASD subjects collected from that site i.e., local imaging markers, while the selected features from in the global model are the global markers candidates that define the ASD subjects in the whole dataset.
In the case of the global model, the input to the RFECV step is and the output is where the size of is such that M ≤ 554. In the case of the site-based model, the input to the RFECV step is 12 (normalized data matrices of each site), and the output is where the size of is such that N is the number of subjects within site k, and M ≤ 544.
Eventually, a global neuro-atlas is created using the whole data set, and a local neuro-atlas is created for each site. We claim that the globla neuro-atlas, as well as, the local neuro-atlases can be used as a guide for future analysis of ASD or ABIDE I dataset.
2.6. ML Classifiers
Having the imaging markers candidates, they should be placed under test to see how good they are at separating the two classes. A set of eight different ML classifiers representing both linear and non-linear hypotheses is selected to test the local, and the global imaging markers candidates. The utilized eight ML classifiers are split into two main divisions: (i) linear classifiers, and (ii) non-linear classifiers. The linear classifiers set comprises LR, LSVM, and passive aggressive. The non-linear classifiers set comprises RF, SVM with radial basis function (SVM-RBF), eXtra Gradient Boost trees (xgboost), Gaussian Naive Bayes (GNB), and neural network (NN) shallow and deep.
To optimize the hyper-parameters of each classifier, the data matrix is reduced accordingly to the results of RFECV algorithm. The data is split five fold. For each of the predefined classifiers, the hyper-parameters and their ranges, where the search will be conducted, are defined in the
Supplementary Material S2, and then a nested for-loop for each classifier. For each hyper-parameter value of that classifier, a five-fold CV is performed, and the results of each fold are saved.
Eventually, the hyper-parameters that corresponds to the maximum CV average score is saved as the optimum parameters. The results of the highest performed classifiers are saved with their hyper-parameter values. The performance metric for each classifier is set to the balanced accuracy score. The detailed algorithm is shown in Algorithm 2.
| Algorithm 2: ML-train with hyper-parameter optimization. |
![Diagnostics 12 00165 i002 Diagnostics 12 00165 i002]() |
2.7. Personalized Diagnosis
We propose a personalized map per subject to show the affected brain regions and to gage the probability of a diagnostic difference when comparing autistic individuals to control. We define a personalized map as a set of scores associated with a set of features denoting the importance of a particular feature in diagnosing a subject as TD or ASD. In a previous work [
13], we created a personalized map for ABIDE I dataset using both fMRI and sMRI features. However, in the proposed work, we are introducing a personalized map for ABIDE I dataset using only sMRI morphological features. The motivation behind creating the personalized map for only the sMRI morphological features is the high performance of the proposed pipeline.
A personalized map is created for each local model. The personalized maps are easily created with classifiers that either associate weights to the input features, such as linear classifiers, or place the input features in a tree schematic that denotes the importance of each feature based on the level of the feature. However, since NN is the used classifier for the local models, it is difficult to determine which input feature contributes the most to the classification decision and which input feature contributes the least.
Local interpretable model-agnostic explanations (LIME) [
75] is a novel explanation technique that explains the prediction of any classifier. The main idea behind LIME is that it builds linear models around the predictions of an opaque model to explain it. LIME is used to explain the classification decisions made by four different local models We have used LIME to explain the decisions of four local models on two random subjects within each of the four sites. Afterward, we visualize the scores representing the contribution of each feature in the classification decision.