Applied Machine Learning in Spiral Breast-CT: Can We Train a Deep Convolutional Neural Network for Automatic, Standardized and Observer Independent Classification of Breast Density?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. BCT Examinations
2.3. Breast Density
2.4. Data Preparation
2.5. dCNN Architecture and Training
2.6. Human Readout “Real-World” Subsets
2.7. Statistical Analyses
3. Results
3.1. Patient Selection and Image Processing
3.1.1. Patient Cohort
3.1.2. Accuracies in Training, Validation and “Real-World” Test Datasets
3.1.3. Human Readout
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno-Aspitia, A.; Advani, P. Current strategies for the prevention of breast cancer. Breast Cancer Targets Ther. 2014, 6, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Plevritis, S.K.; Munoz, D.; Kurian, A.W.; Stout, N.K.; Alagoz, O.; Near, A.M.; Lee, S.J.; Broek, J.J.V.D.; Huang, X.; Schechter, C.B.; et al. Association of Screening and Treatment with Breast Cancer Mortality by Molecular Subtype in US Women, 2000–2012. JAMA J. Am. Med Assoc. 2018, 319, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Boyd, N.F.; Martin, L.J.; Sun, L.; Guo, H.; Chiarelli, A.; Hislop, G.; Yaffe, M.; Minkin, S. Body Size, Mammographic Density, and Breast Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2006, 15, 2086–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamangar, F.; Dores, G.M.; Anderson, W.F. Patterns of Cancer Incidence, Mortality, and Prevalence Across Five Continents: Defining Priorities to Reduce Cancer Disparities in Different Geographic Regions of the World. J. Clin. Oncol. 2006, 24, 2137–2150. [Google Scholar] [CrossRef]
- Lam, P.B.; Vacek, P.M.; Geller, B.M.; Muss, H.B. The association of increased weight, body mass index, and tissue density with the risk of breast carcinoma in Vermont. Cancer 2000, 89, 369–375. [Google Scholar] [CrossRef]
- Burton, A.; Maskarinec, G.; Perez-Gomez, B.; Vachon, C.; Miao, H.; Lajous, M.; López-Ridaura, R.; Rice, M.; Pereira, A.; Garmendia, M.L.; et al. Mammographic density and ageing: A collaborative pooled analysis of cross-sectional data from 22 countries worldwide. PLoS Med. 2017, 14, e1002335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spak, D.; Plaxco, J.; Santiago, L.; Dryden, M.; Dogan, B. BI-RADS ® fifth edition: A summary of changes. Diagn. Interv. Imaging 2017, 98, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, A.B. Redefining the sensitivity of screening mammography: A review. Am. J. Surg. 2019, 218, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Kolb, T.M.; Lichy, J.; Newhouse, J.H. Comparison of the Performance of Screening Mammography, Physical Examination, and Breast US and Evaluation of Factors that Influence Them: An Analysis of 27,825 Patient Evaluations. Radiology 2002, 225, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Wöckel, A.; Festl, J.; Stüber, T.; Brust, K.; Krockenberger, M.; Heuschmann, P.U.; Jírů-Hillmann, S.; Albert, U.-S.; Budach, W.; Follmann, M.; et al. Interdisciplinary Screening, Diagnosis, Therapy and Follow-up of Breast Cancer. Guideline of the DGGG and the DKG (S3-Level, AWMF Registry Number 032/045OL, December 2017)—Part 2 with Recommendations for the Therapy of Primary, Recurrent and Advanced Breast Cancer. Geburtshilfe Frauenheilkd. 2018, 78, 1056–1088. [Google Scholar] [CrossRef] [Green Version]
- Berger, N.; Marcon, M.; Frauenfelder, T.; Boss, A. Dedicated Spiral Breast Computed Tomography with a Single Photon-Counting Detector. Investig. Radiol. 2020, 55, 68–72. [Google Scholar] [CrossRef]
- Berger, N.; Marcon, M.; Saltybaeva, N.; Kalender, W.A.; Alkadhi, H.; Frauenfelder, T.; Boss, A. Dedicated Breast Computed Tomography with a Photon-Counting Detector: Initial Results of Clinical In Vivo Imaging. Investig. Radiol. 2019, 54, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Wienbeck, S.; Fischer, U.; Luftner-Nagel, S.; Lotz, J.; Uhlig, J. Contrast-enhanced cone-beam breast-CT (CBBCT): Clinical performance compared to mammography and MRI. Eur. Radiol. 2018, 28, 3731–3741. [Google Scholar] [CrossRef]
- Li, H.; Yin, L.; He, N.; Han, P.; Zhu, Y.; Ma, Y.; Liu, A.; Lu, H.; Gao, Z.; Liu, P.; et al. Comparison of comfort between cone beam breast computed tomography and digital mammography. Eur. J. Radiol. 2019, 120, 108674. [Google Scholar] [CrossRef]
- Shim, S.; Saltybaeva, N.; Berger, N.; Marcon, M.; Alkadhi, H.; Boss, A. Lesion Detectability and Radiation Dose in Spiral Breast CT With Photon-Counting Detector Technology. Investig. Radiol. 2020, 55, 515–523. [Google Scholar] [CrossRef]
- Wienbeck, S.; Uhlig, J.; Luftner-Nagel, S.; Zapf, A.; Surov, A.; von Fintel, E.; Stahnke, V.; Lotz, J.; Fischer, U. The role of cone-beam breast-CT for breast cancer detection relative to breast density. Eur. Radiol. 2017, 27, 5185–5195. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, Y.; Liu, A.; Yin, L.; Han, P.; Li, H.; Zhang, X.; Ye, Z. A Reliability Comparison of Cone-Beam Breast Computed Tomography and Mammography: Breast Density Assessment Referring to the Fifth Edition of the BI-RADS Atlas. Acad. Radiol. 2019, 26, 752–759. [Google Scholar] [CrossRef]
- Wieler, J.; Berger, N.; Frauenfelder, T.; Marcon, M.; Boss, A. Breast density in dedicated breast computed tomography. Medicine 2021, 100, e25844. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep Learning in Mammography. Investig. Radiol. 2017, 52, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Ciritsis, A.; Rossi, C.; De Martini, I.; Eberhard, M.; Marcon, M.; Becker, A.S.; Berger, N.; Boss, A. Determination of mammographic breast density using a deep convolutional neural network. Br. J. Radiol. 2018, 92, 20180691. [Google Scholar] [CrossRef] [PubMed]
- Ciritsis, A.; Rossi, C.; Eberhard, M.; Marcon, M.; Becker, A.; Boss, A. Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making. Eur. Radiol. 2019, 29, 5458–5468. [Google Scholar] [CrossRef]
- Saffari, N.; Rashwan, H.; Abdel-Nasser, M.; Singh, V.K.; Arenas, M.; Mangina, E.; Herrera, B.; Puig, D. Fully Automated Breast Density Segmentation and Classification Using Deep Learning. Diagnostics 2020, 10, 988. [Google Scholar] [CrossRef] [PubMed]
- Kundel, H.L.; Polansky, M. Measurement of Observer Agreement. Radiology 2003, 228, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. Weighted kappa: Nominal scale agreement provision for scaled disagreement or partial credit. Psychol. Bull. 1968, 70, 213–220. [Google Scholar] [CrossRef]
- Delong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Rutter, D.R.; Calnan, M.; Vaile, M.S.; Field, S.; Wade, K.A. Discomfort and pain during mammography: Description, prediction, and prevention. BMJ 1992, 305, 443–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whelehan, P.; Evans, A.; Wells, M.; MacGillivray, S. The effect of mammography pain on repeat participation in breast cancer screening: A systematic review. Breast 2013, 22, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Stomper, P.C.; D’Souza, D.J.; DiNitto, P.A.; Arredondo, M.A. Analysis of parenchymal density on mammograms in 1353 women 25–79 years old. Am. J. Roentgenol. 1996, 167, 1261–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekpo, E.U.; Ujong, U.P.; Mello-Thoms, C.; McEntee, M.F. Assessment of Interradiologist Agreement Regarding Mammographic Breast Density Classification Using the Fifth Edition of the BI-RADS Atlas. Am. J. Roentgenol. 2016, 206, 1119–1123. [Google Scholar] [CrossRef]
- Winkel, R.R.; Von Euler-Chelpin, M.; Nielsen, M.; Diao, P.; Nielsen, M.B.; Uldall, W.Y.; Vejborg, I. Inter-observer agreement according to three methods of evaluating mammographic density and parenchymal pattern in a case control study: Impact on relative risk of breast cancer. BMC Cancer 2015, 15, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]

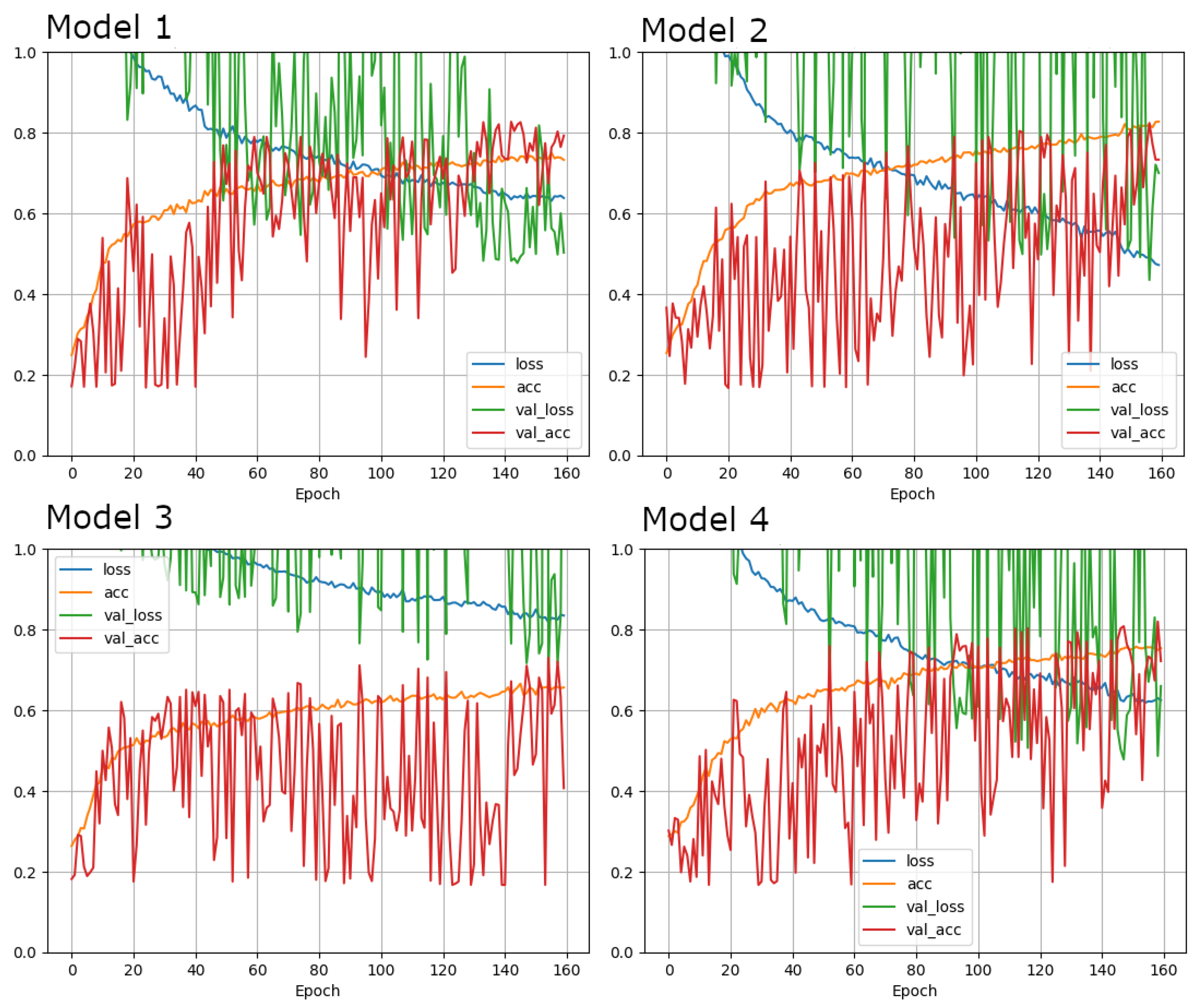

| Model 1 | Model 2 | Model 3 | Model 4 | |
|---|---|---|---|---|
| Architecture | brayZNet | brayZNet | brayZNet | brayZNet |
| Learning rate | 1 × 10−5 | 1 × 10−5 | 1 × 10−5 | 1 × 10−5 |
| Loss function | Cross entropy | Cross entropy | Cross entropy | Cross entropy |
| Optimizer | Adam | Adam | SGD | Adam |
| Augmentation | {‘zooming’: 0.1, ‘rotation’: 45.0, ‘horizontal_shift’: 0.1, ‘vertical_shift’: 0.1, ‘brightness’: 0.0} | {‘zooming’: 0.1, ‘rotation’: 45.0, ‘horizontal_shift’: 0.1, ‘vertical_shift’: 0.1, ‘brightness’: 0.0} | {‘zooming’: 0.1, ‘rotation’: 45.0, ‘horizontal_shift’: 0.1, ‘vertical_shift’: 0.1, ‘brightness’: 0.0} | {‘zooming’: 0.1, ‘rotation’: 45.0, ‘horizontal_shift’: 0.1, ‘vertical_shift’: 0.1, ‘brightness’: 0.0} |
| Epochs | 160 | 160 | 160 | 160 |
| Batch size | 8 | 8 | 8 | 8 |
| Dropout | 0.5 | 0.5 | 0.5 | 0.5 |
| Input Shape | [512, 512, 1] | [256, 256, 1] | [512, 512, 1] | [512, 512, 1] |
| Cropping | None | None | None | [0.12826739057573872, 0.8474049572056288, 0.200998651126856, 0.8363919170216573] |
| Dense layers | 2 | 2 | 2 | 2 |
| Units in layer | 128 | 128 | 128 | 128 |
| Regularization | l1 = 1 × 10−6, l2 = 1 × 10−6 | l1 = 1 × 10−6, l2 = 1 × 10−6 | l1 = 1 × 10−6, l2 = 1 × 10−6 | l1 = 1 × 10−6, l2 = 1 × 10−6 |
| Test accuracy | 0.8041666746139526 | 0.8583333492279053 | 0.7354166507720947 | 0.8020833134651184 |
| Density Level | Ultrasound (US) | Reason for US Examination | |||
|---|---|---|---|---|---|
| Yes | No | Density | Other | ||
| A | 58 | 14 | 44 | 0 | 14 |
| B | 118 | 64 | 54 | 28 | 36 |
| C | 83 | 80 | 3 | 57 | 23 |
| D | 58 | 56 | 2 | 43 | 13 |
| Total n | 317 | 214 | 103 | 128 | 86 |
| Predicted Density Level | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 3 | Model 4 | ||||||||||||||
| A | B | C | D | A | B | C | D | A | B | C | D | A | B | C | D | ||
| Density level (ground truth) | A | 91 | 15 | 0 | 0 | 90 | 15 | 1 | 0 | 93 | 13 | 0 | 0 | 63 | 43 | 0 | 0 |
| B | 19 | 131 | 23 | 0 | 12 | 142 | 19 | 0 | 31 | 129 | 10 | 3 | 4 | 154 | 15 | 0 | |
| C | 0 | 25 | 101 | 2 | 0 | 11 | 112 | 5 | 1 | 38 | 88 | 1 | 0 | 20 | 106 | 2 | |
| D | 0 | 0 | 10 | 63 | 0 | 1 | 4 | 68 | 0 | 1 | 29 | 43 | 0 | 0 | 11 | 62 | |
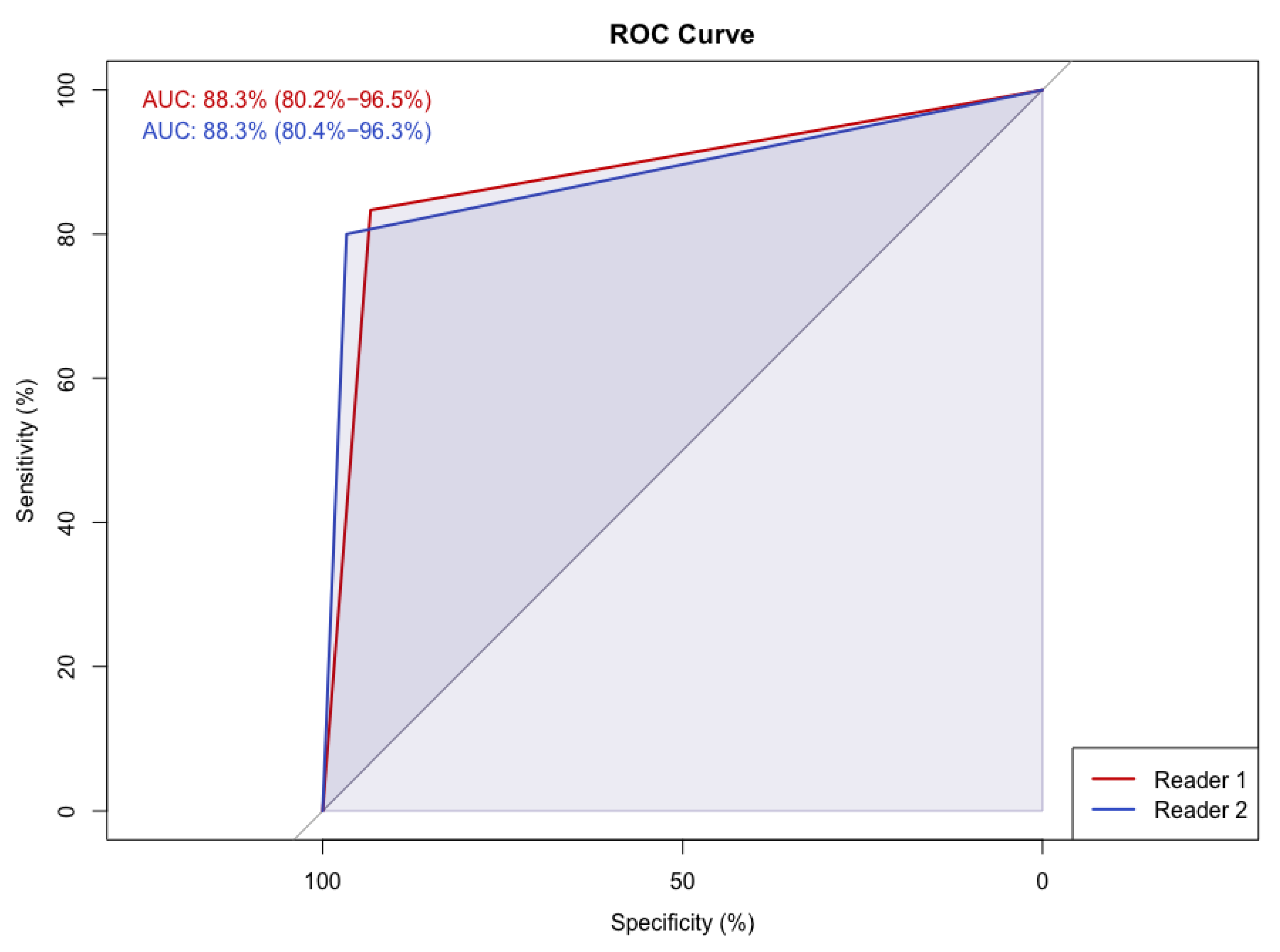
| dCNN | Reader 1 | Reader 2 | ||
|---|---|---|---|---|
| A | 14 | 15 | 16 | Low density |
| B | 16 | 18 | 19 | |
| C | 15 | 17 | 10 | High density |
| D | 15 | 10 | 15 |
| Ground-Truth | dCNN | Reader 1 | Reader 2 | |
|---|---|---|---|---|
| Ground-Truth | 0.84 | 0.87 | 0.82 | |
| dCNN | 0.71 | 0.73 | ||
| Reader 1 | 0.73 | |||
| Reader 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landsmann, A.; Wieler, J.; Hejduk, P.; Ciritsis, A.; Borkowski, K.; Rossi, C.; Boss, A. Applied Machine Learning in Spiral Breast-CT: Can We Train a Deep Convolutional Neural Network for Automatic, Standardized and Observer Independent Classification of Breast Density? Diagnostics 2022, 12, 181. https://doi.org/10.3390/diagnostics12010181
Landsmann A, Wieler J, Hejduk P, Ciritsis A, Borkowski K, Rossi C, Boss A. Applied Machine Learning in Spiral Breast-CT: Can We Train a Deep Convolutional Neural Network for Automatic, Standardized and Observer Independent Classification of Breast Density? Diagnostics. 2022; 12(1):181. https://doi.org/10.3390/diagnostics12010181
Chicago/Turabian StyleLandsmann, Anna, Jann Wieler, Patryk Hejduk, Alexander Ciritsis, Karol Borkowski, Cristina Rossi, and Andreas Boss. 2022. "Applied Machine Learning in Spiral Breast-CT: Can We Train a Deep Convolutional Neural Network for Automatic, Standardized and Observer Independent Classification of Breast Density?" Diagnostics 12, no. 1: 181. https://doi.org/10.3390/diagnostics12010181
APA StyleLandsmann, A., Wieler, J., Hejduk, P., Ciritsis, A., Borkowski, K., Rossi, C., & Boss, A. (2022). Applied Machine Learning in Spiral Breast-CT: Can We Train a Deep Convolutional Neural Network for Automatic, Standardized and Observer Independent Classification of Breast Density? Diagnostics, 12(1), 181. https://doi.org/10.3390/diagnostics12010181






