A Machine Learning Ensemble Based on Radiomics to Predict BI-RADS Category and Reduce the Biopsy Rate of Ultrasound-Detected Suspicious Breast Masses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population and Image Sets
2.2. Radiomic-Based Machine Learning Modelling
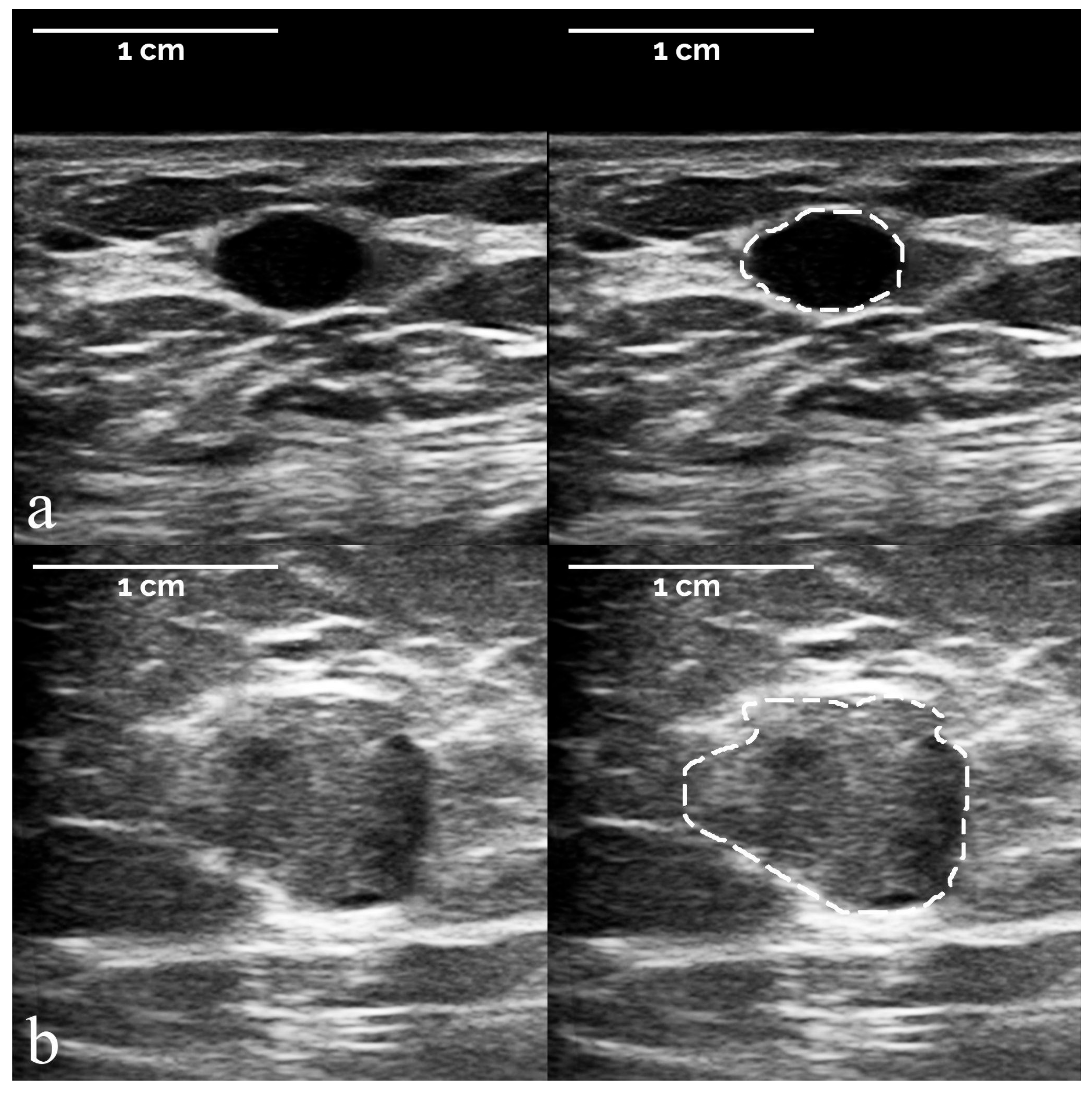
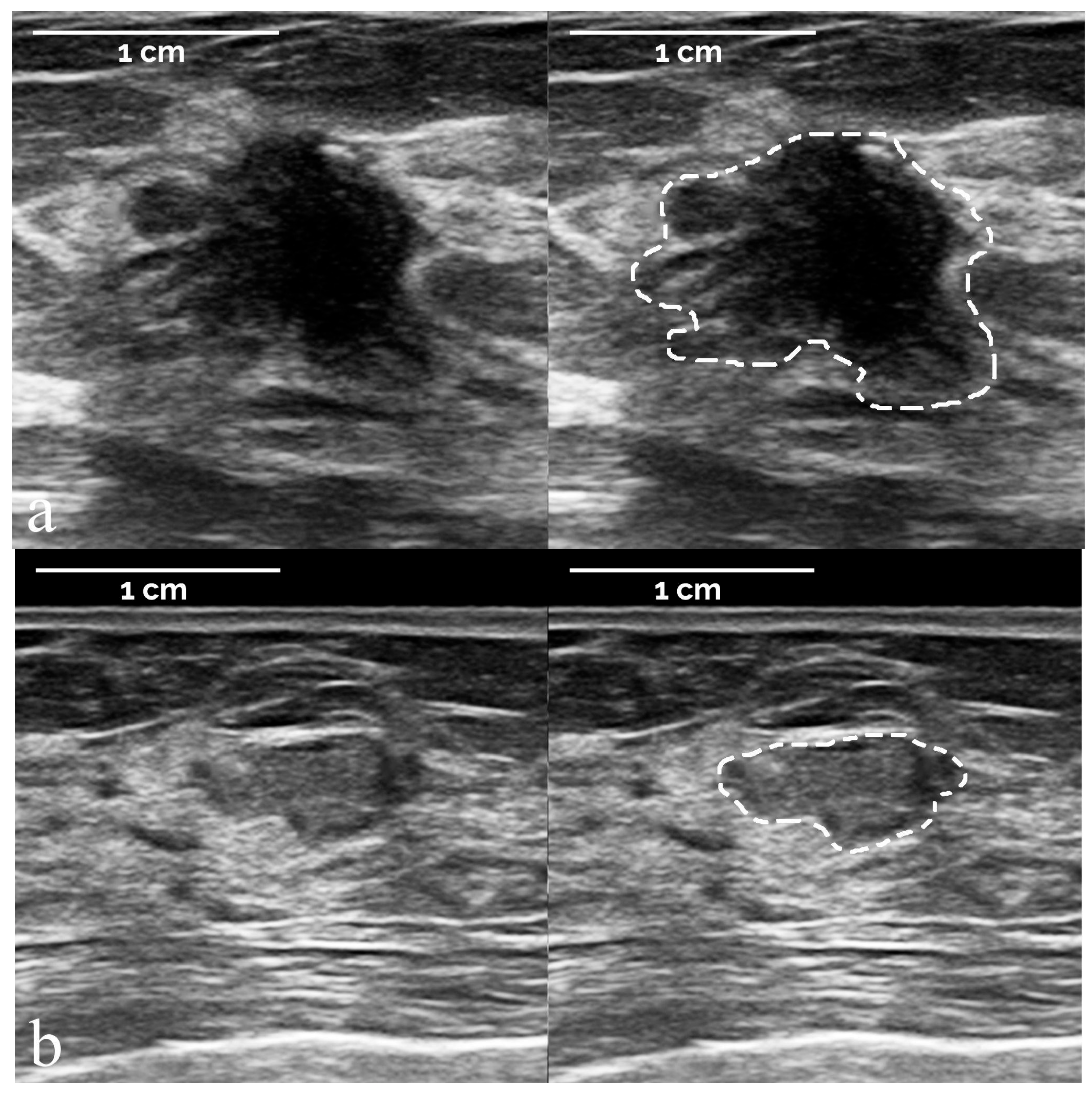
- The segmentation of suspicious masses on all 821 images was performed manually by a board-certified radiologist with 34 years of experience in breast imaging, using the TRACE4 segmentation tool. The same radiologist (at a time distance of 8 weeks) and a second board-certified radiologist with 7 years of experience independently segmented the masses on a random subsample of 50 images from the training dataset, fully blinded to histopathology and other segmentations.
- The preprocessing of image intensities within the segmented ROI included resampling to isotropic voxel spacing, using a downsampling scheme by considering an image slice thickness equal to pixel spacing, and intensity discretization using a fixed number of 64 bins.
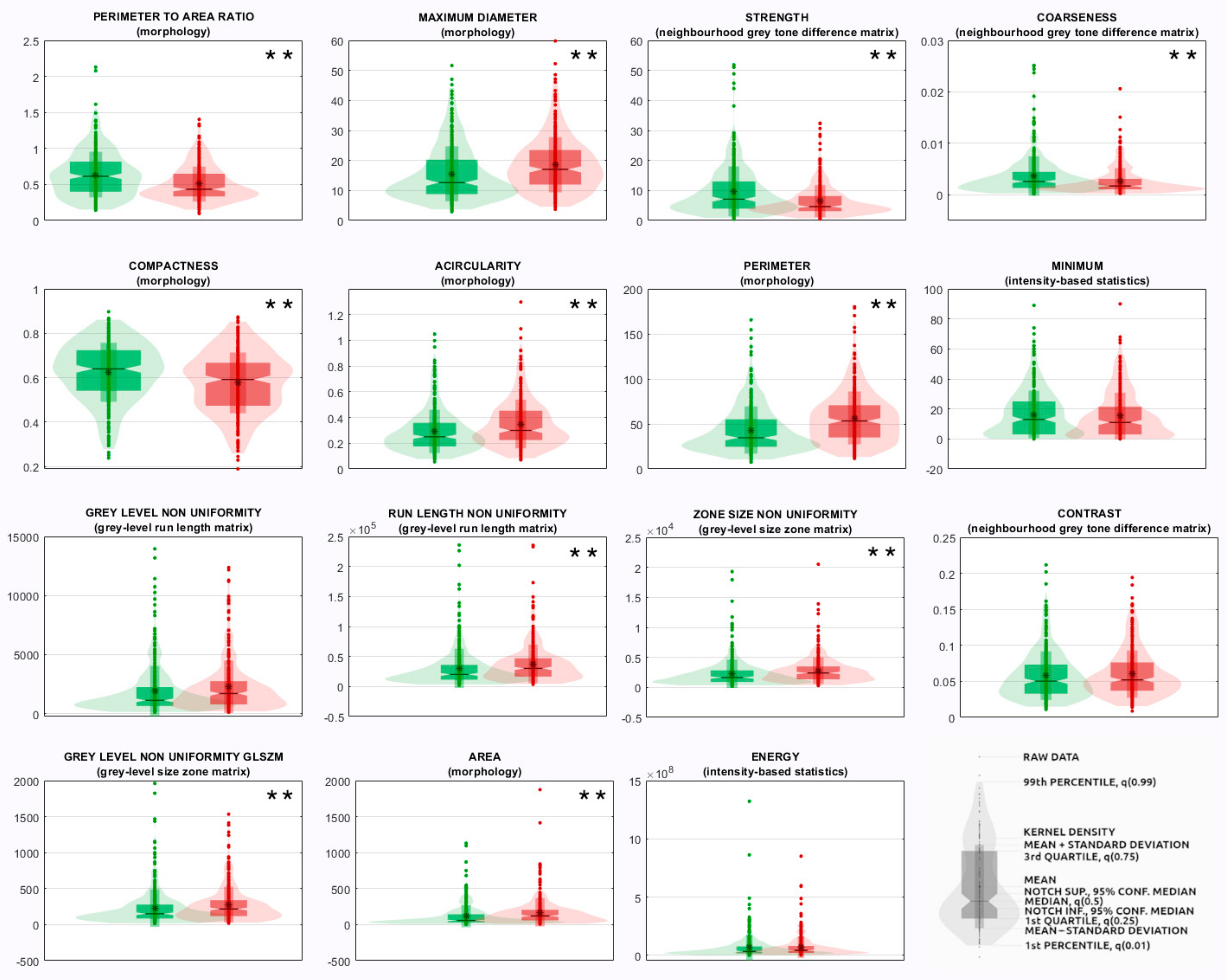
- The radiomics features measured from the segmented ROI were 107 quantitative descriptors and belonged to different families: morphology, intensity-based statistics, intensity histogram, grey-level co-occurrence matrix (GLCM), grey-level run length matrix (GLRLM), grey-level size zone matrix (GLSZM), neighborhood grey tone difference matrix (NGTDM), grey-level distance zone matrix (GLDZM), and neighboring grey-level dependence matrix (NGLDM). Their definition, computation, and nomenclature are compliant with the IBSI guidelines, except for the features of the family morphology, originally designed for 3D images, which were replaced with ten 2D equivalent features (e.g., 3D features volume and surface were replaced with 2D features area and perimeter, respectively). Radiomic features were selected as those showing an intraclass correlation coefficient >0.75 among the two intra-observer and inter-observer segmentations on the random subsample of images described in point (1), since according to the 95% confidence interval of the intraclass correlation coefficient estimate, values lower than 0.5, between 0.5 and 0.75, between 0.75 and 0.9, and higher than 0.9 are indicative of poor, moderate, good, and excellent reliability, respectively [14]. Steps from (2) to (3) were performed using the TRACE4 Radiomics tool. Radiomic features were reported by TRACE4 according to IBSI standards.
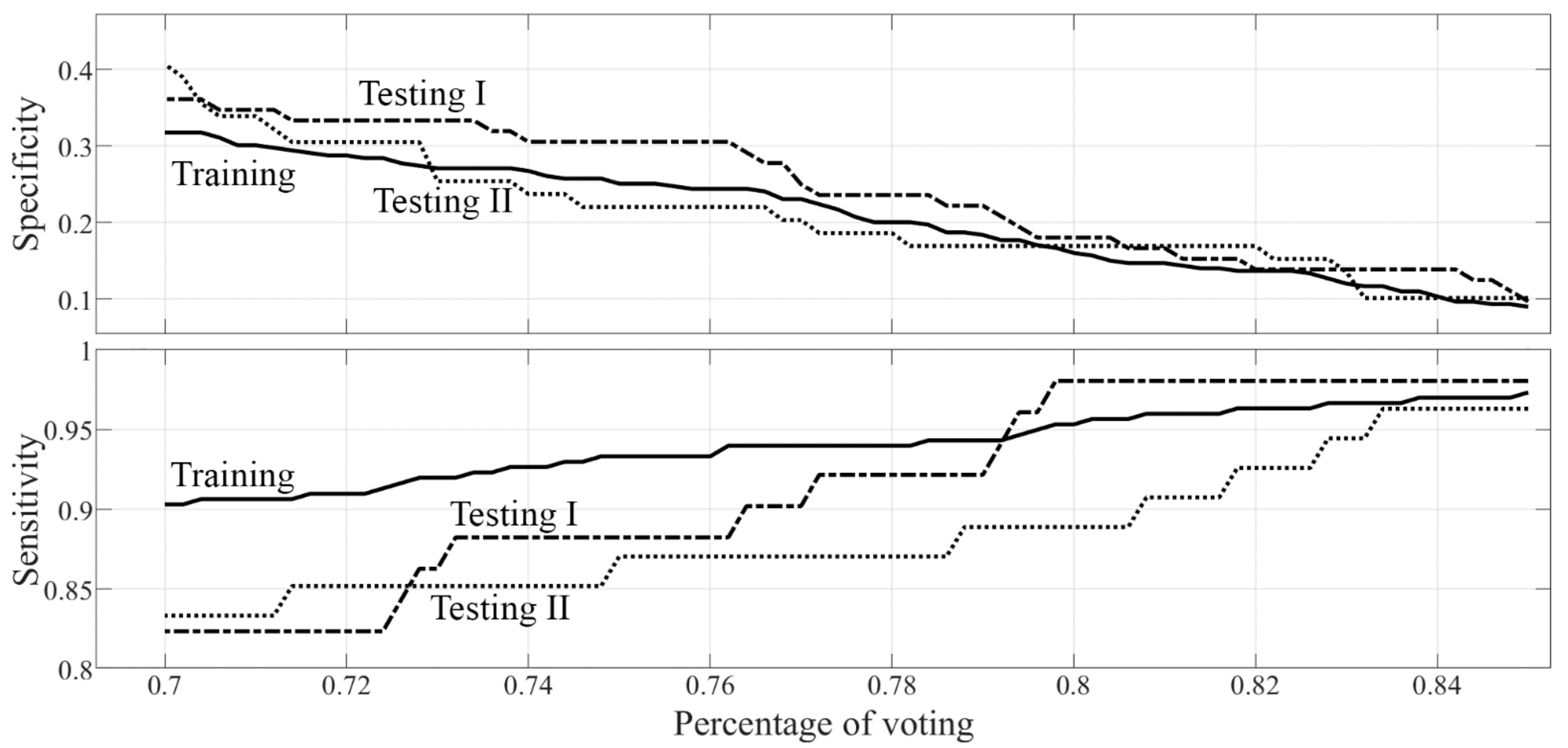
- Three different models of machine learning classifiers were trained, validated, and tested, for the binary classification task of interest (malignant versus benign discrimination), based on supervised learning, using histopathology as a reference standard. For each model, a nested k-fold cross-validation method was used (k = 10, 8 folds for training, 1 fold for tuning, 1 fold for hold-out testing, random sampling). The first model consisted of 3 ensembles of 100 random forest classifiers combined with Gini index with majority-vote rule; the second model consisted of 3 ensembles of 100 support vector machines (linear kernel) combined with principal components analysis and Fisher discriminant ratio with majority-vote rule; the third ensemble consisted of 3 ensembles of 100 k-nearest neighbor classifiers combined with principal components analysis and Fisher discriminant ratio with majority-vote rule. Data for the ensemble learning set were selected by using 100 baggings based on random sampling without replacement (80% data for training, 10% data for tuning, 10% data for internal testing). Each classifier belonging to the same ensemble was tested internally on datasets that can not have data samples in common. Classifiers belonging to different ensembles were tested on datasets that can have samples in common. The performances of the 3 models were measured across the different folds (k = 10) in terms of sensitivity, specificity, area under the receiver operating characteristic curve (ROC-AUC), positive predictive value (PPV), negative predictive value (NPV), and corresponding 95% confidence intervals (CI). The model with the best performance according to ROC-AUC was chosen as the best classification model for the binary task of interest (malignant versus benign discrimination).
2.3. BI-RADS Diagnostic Categories Classification
2.4. Statistical Analysis
3. Results
3.1. Study Population and Image Sets
3.2. Radiomic-Based Machine Learning Modelling
3.3. BI-RADS Diagnostic Categories Classification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Evans, A.; Trimboli, R.M.; Athanasiou, A.; Balleyguier, C.; Baltzer, P.A.; Bick, U.; Camps Herrero, J.; Clauser, P.; Colin, C.; Cornford, E.; et al. Breast ultrasound: Recommendations for information to women and referring physicians by the European Society of Breast Imaging. Insights Imaging 2018, 9, 449–461. [Google Scholar] [CrossRef] [Green Version]
- Bick, U.; Trimboli, R.M.; Athanasiou, A.; Balleyguier, C.; Baltzer, P.A.T.; Bernathova, M.; Borbély, K.; Brkljacic, B.; Carbonaro, L.A.; Clauser, P.; et al. Image-guided breast biopsy and localisation: Recommendations for information to women and referring physicians by the European Society of Breast Imaging. Insights Imaging 2020, 11, 12. [Google Scholar] [CrossRef]
- Tomkovich, K.R. Interventional Radiology in the Diagnosis and Treatment of Diseases of the Breast: A Historical Review and Future Perspective Based on Currently Available Techniques. Am. J. Roentgenol. 2014, 203, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Biganzoli, L.; Marotti, L.; Hart, C.D.; Cataliotti, L.; Cutuli, B.; Kühn, T.; Mansel, R.E.; Ponti, A.; Poortmans, P.; Regitnig, P.; et al. Quality indicators in breast cancer care: An update from the EUSOMA working group. Eur. J. Cancer 2017, 86, 59–81. [Google Scholar] [CrossRef]
- D’Orsi, C.J.; Sickles, E.A.; Mendelson, E.B.; Morris, E.A. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System, 5th ed.; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-H.; Park, H.; Ko, E.S. Radiomics in Breast Imaging from Techniques to Clinical Applications: A Review. Korean J. Radiol. 2020, 21, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, A.; Daimiel Naranjo, I.; Lo Gullo, R.; Rossi Saccarelli, C.; Pinker, K. AI-enhanced breast imaging: Where are we and where are we heading? Eur. J. Radiol. 2021, 142, 109882. [Google Scholar] [CrossRef]
- Hu, Q.; Giger, M.L. Clinical Artificial Intelligence Applications: Breast Imaging. Radiol. Clin. N. Am. 2021, 59, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Stavros, A.T.; Thickman, D.; Rapp, C.L.; Dennis, M.A.; Parker, S.H.; Sisney, G.A. Solid breast nodules: Use of sonography to distinguish between benign and malignant lesions. Radiology 1995, 196, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- TRACE4. Available online: http://www.deeptracetech.com/temp/TechnicalSheet__TRACE4.pdf (accessed on 7 January 2022).
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Meani, F.; Di Lascio, S.; Wandschneider, W.; Montagna, G.; Vitale, V.; Zehbe, S.; Harder, Y.; Parvex, S.L.; Spina, P.; Canonica, C.; et al. Granular cell tumor of the breast: A multidisciplinary challenge. Crit. Rev. Oncol. Hematol. 2019, 144, 102828. [Google Scholar] [CrossRef]
- Van Breest Smallenburg, V.; Nederend, J.; Voogd, A.C.; Coebergh, J.W.W.; van Beek, M.; Jansen, F.H.; Louwman, W.J.; Duijm, L.E.M. Trends in breast biopsies for abnormalities detected at screening mammography: A population-based study in the Netherlands. Br. J. Cancer 2013, 109, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Allison, K.H.; Abraham, L.A.; Weaver, D.L.; Tosteson, A.N.A.; Nelson, H.D.; Onega, T.; Geller, B.M.; Kerlikowske, K.; Carney, P.A.; Ichikawa, L.E.; et al. Trends in breast biopsy pathology diagnoses among women undergoing mammography in the United States: A report from the Breast Cancer Surveillance Consortium. Cancer 2015, 121, 1369–1378. [Google Scholar] [CrossRef] [Green Version]
- Mendelson, E.B.; Böhm-Vélez, M.; Berg, W.A. ACR BI-RADS® Ultrasound. In ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System; American College of Radiology: Reston, VA, USA, 2013. [Google Scholar]
- Fleury, E.; Marcomini, K. Performance of machine learning software to classify breast lesions using BI-RADS radiomic features on ultrasound images. Eur. Radiol. Exp. 2019, 3, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Islami, F.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer in Women: Burden and Trends. Cancer Epidemiol. Biomark. Prev. 2017, 26, 444–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Vecchia, C.; Carioli, G. The epidemiology of breast cancer, a summary overview. Epidemiol. Biostat. Public Health 2018, 15, e12853-1–e12853-2. [Google Scholar] [CrossRef]
- Flobbe, K.; Bosch, A.M.; Kessels, A.G.H.; Beets, G.L.; Nelemans, P.J.; von Meyenfeldt, M.F.; van Engelshoven, J.M.A. The Additional Diagnostic Value of Ultrasonography in the Diagnosis of Breast Cancer. Arch. Intern. Med. 2003, 163, 1194. [Google Scholar] [CrossRef] [Green Version]
- McCavert, M.; O’Donnell, M.E.; Aroori, S.; Badger, S.A.; Sharif, M.A.; Crothers, J.G.; Spence, R.A.J. Ultrasound is a useful adjunct to mammography in the assessment of breast tumours in all patients. Int. J. Clin. Pract. 2009, 63, 1589–1594. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Lu, G.; Qin, B.; Fei, B. Ultrasound Imaging Technologies for Breast Cancer Detection and Management: A Review. Ultrasound Med. Biol. 2018, 44, 37–70. [Google Scholar] [CrossRef]
- Spick, C.; Baltzer, P.A.T. Diagnostic Utility of Second-Look US for Breast Lesions Identified at MR Imaging: Systematic Review and Meta-Analysis. Radiology 2014, 273, 401–409. [Google Scholar] [CrossRef]
- Mango, V.L.; Sun, M.; Wynn, R.T.; Ha, R. Should We Ignore, Follow, or Biopsy? Impact of Artificial Intelligence Decision Support on Breast Ultrasound Lesion Assessment. Am. J. Roentgenol. 2020, 214, 1445–1452. [Google Scholar] [CrossRef]
- Baltzer, P.A.T.; Sardanelli, F. The Mantra about Low Specificity of Breast MRI. In Breast MRI for High-Risk Screening; Sardanelli, F., Podo, F., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 11–21. [Google Scholar]
- Andreu, F.J.; Sáez, A.; Sentís, M.; Rey, M.; Fernández, S.; Dinarès, C.; Tortajada, L.; Ganau, S.; Palomar, G. Breast core biopsy reporting categories—An internal validation in a series of 3054 consecutive lesions. Breast 2007, 16, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Youk, J.H.; Kim, E.-K.; Kim, M.J.; Oh, K.K. Sonographically Guided 14-Gauge Core Needle Biopsy of Breast Masses: A Review of 2420 Cases with Long-Term Follow-Up. Am. J. Roentgenol. 2008, 190, 202–207. [Google Scholar] [CrossRef]
- El-Sayed, M.E.; Rakha, E.A.; Reed, J.; Lee, A.H.; Evans, A.J.; Ellis, I.O. Audit of performance of needle core biopsy diagnoses of screen detected breast lesions. Eur. J. Cancer 2008, 44, 2580–2586. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.; Han, K.; Kim, M.J.; Moon, H.J.; Yoon, J.H.; Park, V.Y.; Kim, E.-K. Annual Trends in Ultrasonography-Guided 14-Gauge Core Needle Biopsy for Breast Lesions. Korean J. Radiol. 2020, 21, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Helbich, T.H.; Matzek, W.; Fuchsjäger, M.H. Stereotactic and ultrasound-guided breast biopsy. Eur. Radiol. 2004, 14, 383–393. [Google Scholar] [CrossRef]
- O’Flynn, E.A.M.; Wilson, A.R.M.; Michell, M.J. Image-guided breast biopsy: State-of-the-art. Clin. Radiol. 2010, 65, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xiao, M.; Liu, H.; Wang, M.; Wang, H.; Zhang, J.; Jiang, Y.; Zhu, Q. Reducing the number of unnecessary biopsies of US-BI-RADS 4a lesions through a deep learning method for residents-in-training: A cross-sectional study. BMJ Open 2020, 10, e035757. [Google Scholar] [CrossRef] [PubMed]
- Barinov, L.; Jairaj, A.; Paster, L.; Hulbert, W.; Mammone, R.; Podilchuk, C. Decision quality support in diagnostic breast ultrasound through Artificial Intelligence. In Proceedings of the 2016 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 3 December 2016; pp. 1–4. [Google Scholar]
- Lee, J.; Kim, S.; Kang, B.J.; Kim, S.H.; Park, G.E. Evaluation of the effect of computer aided diagnosis system on breast ultrasound for inexperienced radiologists in describing and determining breast lesions. Med. Ultrason. 2019, 21, 239–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Carmen, M.G.; Halpern, E.F.; Kopans, D.B.; Moy, B.; Moore, R.H.; Goss, P.E.; Hughes, K.S. Mammographic Breast Density and Race. Am. J. Roentgenol. 2007, 188, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.-M.; Kim, E.H. Breast Density and Risk of Breast Cancer in Asian Women: A Meta-analysis of Observational Studies. J. Prev. Med. Public Health 2016, 49, 367–375. [Google Scholar] [CrossRef] [Green Version]
- Rajaram, N.; Mariapun, S.; Eriksson, M.; Tapia, J.; Kwan, P.Y.; Ho, W.K.; Harun, F.; Rahmat, K.; Czene, K.; Taib, N.A.M.; et al. Differences in mammographic density between Asian and Caucasian populations: A comparative analysis. Breast Cancer Res. Treat. 2017, 161, 353–362. [Google Scholar] [CrossRef]
| Malignant or Benign | Histopathology Type | Number | Percentage |
|---|---|---|---|
| Benign | Fibroadenoma | 146 | 34.0% |
| Sclerosing lesions/adenosis | 64 | 14.9% | |
| Normal breast tissue | 38 | 8.8% | |
| Inflammatory lesions | 36 | 8.4% | |
| Papilloma (no atypia) | 27 | 6.3% | |
| Cysts, ductal ectasia, or seromas | 37 | 8.6% | |
| Usual ductal hyperplasia | 17 | 3.9% | |
| Atypical ductal hyperplasia | 8 | 1.9% | |
| Fibroadenomatoid changes | 23 | 5.3% | |
| Other benign findings | 34 | 7.9% | |
| Total | 430 | 100% | |
| Malignant | Invasive ductal carcinoma | 304 | 75.2% |
| Invasive lobular carcinoma | 42 | 10.4% | |
| Ductal carcinoma in situ | 19 | 4.7% | |
| Other malignancies originating from breast tissues | 35 | 8.7% | |
| Other malignancies (metastases from non breast tissues) | 4 | 1.0% | |
| Total | 404 | 100% |
| Dataset | US System | US Images | Total Lesions | Mean Pixel Size (Range) (mm) | Malignant Lesions | Mean Pixel Size (Range) (mm) | Benign Lesions | Mean Pixel Size (Range) (mm) |
|---|---|---|---|---|---|---|---|---|
| Training and internaltesting | Esaote MyLab 6100 | 273 | 277 | 0.098 | 156 | 0.103 | 121 | 0.092 |
| (0.046–0.154) | (0.046–0.154) | (0.046–0.139) | ||||||
| Esaote MyLab 6150 | 311 | 318 | 0.091 | 142 | 0.095 | 176 | 0.088 | |
| (0.046–0.123) | (0.046–0.123) | (0.046–0.123) | ||||||
| Esaote MyLab 6440 | 2 | 3 | 0.068 | 1 | 0.068 | 2 | 0.068 | |
| (0.068–0.068) | (0.068–0.068) | (0.068–0.068) | ||||||
| Testing set I | Esaote MyLab 6100 | 59 | 59 | 0.091 | 20 | 0.094 | 39 | 0.090 |
| (0.046–0.109) | (0.062–0.108) | (0.046–0.108) | ||||||
| Esaote MyLab 6150 | 63 | 63 | 0.097 | 31 | 0.101 | 32 | 0.092 | |
| (0.048–0.139) | (0.048–0.139) | (0.062–0.123) | ||||||
| Esaote MyLab 7340002 | 1 | 1 | 0.106 | 0 | – | 1 | 0.106 | |
| (0.106–0.106) | (0.106–0.106) | |||||||
| Testing set II | Samsung RS80A | 86 | 86 | 0.065 | 44 | 0.068 | 42 | 0.062 |
| (0.040–0.110) | (0.050–0.110) | (0.040–0.090) | ||||||
| Siemens Healthineers Acuson Juniper | 26 | 27 | 0.067 | 10 | 0.069 | 17 | 0.066 | |
| (0.030–0.080) | (0.060–0.070) | (0.030–0.080) |
| Performance Metric | Testing Set I | Testing Set II |
|---|---|---|
| SVM sensitivity (95% CI) | 98.0% (89.6%–99.9%) | 94.4% (84.6%–98.8%) |
| SVM NPV (95% CI) | 92.9% (66.1%–99.8%) | 75.0% (42.8%–94.5%) |
| SVM PPV (95% CI) | 45.9% ** (36.3%–55.7%) | 50.5% ** (40.4%–60.6%) |
| Radiologists’ PPV | 41.5% ** (32.7%–50.7%) | 47.8% ** (38.3%–57.4) |
| SVM specificity (95% CI) | 18.1% ** (10.0%–28.9%) | 15.3% ** (7.2%–27.0%) |
| Radiologists’ specificity | 0.0% | 0.0% |
| Rank | Feature Family | Feature Name |
|---|---|---|
| 1 | Morphology | Perimeter-to-area ratio ** |
| 2 | Morphology | Maximum diameter ** |
| 3 | Morphology | Compactness ** |
| 4 | Morphology | Acircularity ** |
| 5 | Morphology | Perimeter ** |
| 6 | Morphology | Area ** |
| 7 | Morphology | Center of mass shift ** |
| 8 | Morphology | Circularity * |
| 9 | Neighborhood grey tone difference matrix | Strength ** |
| 10 | Neighborhood grey tone difference matrix | Coarseness ** |
| 11 | Neighborhood grey tone difference matrix | Contrast |
| 12 | Neighborhood grey tone difference matrix | Busyness * |
| 13 | Grey-level size zone matrix | Zone size non-uniformity ** |
| 14 | Grey-level size zone matrix | Grey-level non-uniformity glszm ** |
| 15 | Neighboring grey-level dependence matrix | Dependence count non-uniformity ** |
| 16 | Neighboring grey-level dependence matrix | Low-dependence low-grey-level emphasis |
| 17 | Grey-level run length matrix | Grey-level non-uniformity |
| 18 | Grey-level run length matrix | Run length non-uniformity |
| 19 | Intensity-based statistics | Minimum |
| 20 | Intensity-based statistics | Energy |
| 21 | Intensity-based statistics | Variance |
| 22 | Intensity-based statistics | Quartile coefficient |
| 23 | Intensity-based statistics | 10th percentile |
| 24 | Intensity histogram | 10th percentile |
| 25 | Grey-level co-occurrence matrix | First measure of information correlation |
| Histopathology Type | BI-RADS 3 (%) | BI-RADS 4 (%) | BI-RADS 5 (%) |
|---|---|---|---|
| Fibroadenoma | 8 (1.3) | 93 (15.6) | 0 (0.0) |
| Sclerosing lesions/adenosis | 10 (1.7) | 37 (6.2) | 2 (0.3) |
| Normal breast tissue | 6 (1.0) | 26 (4.3) | 1 (0.2) |
| Inflammatory lesions | 2 (0.3) | 26 (4.3) | 0 (0.0) |
| Papilloma (no atypia) | 1 (0.2) | 17 (2.8) | 0 (0.0) |
| Cysts, ductal ectasia, or seromas | 10 (1.7) | 13 (2.2) | 0 (0.0) |
| Usual ductal hyperplasia | 2 (0.3) | 9 (1.5) | 0 (0.0) |
| Atypical ductal hyperplasia | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Fibroadenomatoid changes | 2 (0.3) | 13 (2.2) | 0 (0.0) |
| Other benign findings | 8 (1.3) | 93 (15.6) | 0 (0.0) |
| Invasive ductal carcinoma | 5 (0.8) | 211 (35.3) | 6 (1.0) |
| Invasive lobular carcinoma | 2 (0.3) | 32 (5.4) | 1 (0.2) |
| Ductal carcinoma in situ | 4 (0.7) | 10 (1.7) | 0 (0.0) |
| Other malignancies originating from breast tissues | 2 (0.3) | 24 (4.0) | 0 (0.0) |
| Other malignancies (metastases from non breast tissues) | 0 (0.0) | 1 (0.2) | 1 (0.2) |
| Histopathology Type | BI-RADS 3 (%) | BI-RADS 4 (%) | BI-RADS 5 (%) |
|---|---|---|---|
| Fibroadenoma | 2 (1.6) | 19 (15.4) | 0 (0.0) |
| Sclerosing lesions/adenosis | 1 (0.8) | 5 (4.1) | 0 (0.0) |
| Normal breast tissue | 0 (0.0) | 3 (2.4) | 0 (0.0) |
| Inflammatory lesions | 2 (1.6) | 4 (3.3) | 0 (0.0) |
| Papilloma (no atypia) | 2 (1.6) | 4 (3.3) | 0 (0.0) |
| Cysts, ductal ectasia, or seromas | 2 (1.6) | 6 (4.9) | 0 (0.0) |
| Usual ductal hyperplasia | 0 (0.0) | 4 (3.3) | 0 (0.0) |
| Atypical ductal hyperplasia | 3 (2.4) | 5 (4.1) | 0 (0.0) |
| Fibroadenomatoid changes | 0 (0.0) | 1 (0.8) | 0 (0.0) |
| Other benign findings | 1 (0.8) | 8 (6.5) | 0 (0.0) |
| Invasive ductal carcinoma | 1 (0.8) | 40 (32.5) | 0 (0.0) |
| Invasive lobular carcinoma | 0 (0.0) | 3 (2.4) | 0 (0.0) |
| Ductal carcinoma in situ | 0 (0.0) | 2 (1.6) | 0 (0.0) |
| Other malignancies originating from breast tissues | 0 (0.0) | 4 (3.3) | 0 (0.0) |
| Other malignancies (metastases from non breast tissues) | 0 (0.0) | 1 (0.8) | 0 (0.0) |
| Histopathology Type | BI-RADS 3 (%) | BI-RADS 4 (%) | BI-RADS 5 (%) |
|---|---|---|---|
| Fibroadenoma | 1 (0.9) | 23 (20.4) | 0 (0.0) |
| Sclerosing lesions/adenosis | 2 (1.8) | 7 (6.2) | 0 (0.0) |
| Normal breast tissue | 0 (0.0) | 2 (1.8) | 0 (0.0) |
| Inflammatory lesions | 1 (0.9) | 1 (0.9) | 0 (0.0) |
| Papilloma (no atypia) | 1 (0.9) | 2 (1.8) | 0 (0.0) |
| Cysts, ductal ectasia, or seromas | 1 (0.9) | 5 (4.4) | 0 (0.0) |
| Usual ductal hyperplasia | 1 (0.9) | 1 (0.9) | 0 (0.0) |
| Atypical ductal hyperplasia | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Fibroadenomatoid changes | 1 (0.9) | 6 (5.3) | 0 (0.0) |
| Other benign findings | 1 (0.9) | 3 (2.7) | 0 (0.0) |
| Invasive ductal carcinoma | 1 (0.0) | 40 (35.4) | 0 (0.0) |
| Invasive lobular carcinoma | 0 (0.0) | 4 (3.5) | 0 (0.0) |
| Ductal carcinoma in situ | 1 (0.9) | 2 (1.8) | 0 (0.0) |
| Other malignancies originating from breast tissues | 1 (0.9) | 4 (3.5) | 0 (0.0) |
| Other malignancies (metastases from non breast tissues) | 0 (0.0) | 1 (0.9) | 0 (0.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Interlenghi, M.; Salvatore, C.; Magni, V.; Caldara, G.; Schiavon, E.; Cozzi, A.; Schiaffino, S.; Carbonaro, L.A.; Castiglioni, I.; Sardanelli, F. A Machine Learning Ensemble Based on Radiomics to Predict BI-RADS Category and Reduce the Biopsy Rate of Ultrasound-Detected Suspicious Breast Masses. Diagnostics 2022, 12, 187. https://doi.org/10.3390/diagnostics12010187
Interlenghi M, Salvatore C, Magni V, Caldara G, Schiavon E, Cozzi A, Schiaffino S, Carbonaro LA, Castiglioni I, Sardanelli F. A Machine Learning Ensemble Based on Radiomics to Predict BI-RADS Category and Reduce the Biopsy Rate of Ultrasound-Detected Suspicious Breast Masses. Diagnostics. 2022; 12(1):187. https://doi.org/10.3390/diagnostics12010187
Chicago/Turabian StyleInterlenghi, Matteo, Christian Salvatore, Veronica Magni, Gabriele Caldara, Elia Schiavon, Andrea Cozzi, Simone Schiaffino, Luca Alessandro Carbonaro, Isabella Castiglioni, and Francesco Sardanelli. 2022. "A Machine Learning Ensemble Based on Radiomics to Predict BI-RADS Category and Reduce the Biopsy Rate of Ultrasound-Detected Suspicious Breast Masses" Diagnostics 12, no. 1: 187. https://doi.org/10.3390/diagnostics12010187







