RT-PCR Detection of SARS-CoV-2 among Individuals from the Upper Silesian Region—Analysis of 108,516 Tests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. RNA Isolation
2.3. RT-PCR Assays
2.4. Identification of SARS-CoV-2 Variants
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Timeline of WHO’s Response to COVID-19. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline?gclid=EAIaIQobChMI1uWCv72c9AIVHQWiAx1vVwuuEAAYASAAEgLjTPD_BwE#event-115 (accessed on 16 November 2021).
- Cao, M.; Zhang, D.; Wang, Y.; Lu, Y.; Zhu, X.; Li, Y.; Xue, H.; Lin, Y.; Zhang, M.; Sun, Y.; et al. Clinical Features of Patients Infected with the 2019 Novel Coronavirus (COVID-19) in Shanghai, China. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Grant, M.C.; Geoghegan, L.; Arbyn, M.; Mohammed, Z.; McGuinness, L.; Clarke, E.L.; Wade, R.G. The Prevalence of Symptoms in 24,410 Adults Infected by the Novel Coronavirus (SARS-CoV-2; COVID-19): A Systematic Review and Meta-Analysis of 148 Studies from 9 Countries. PLoS ONE 2020, 15, e0234765. [Google Scholar] [CrossRef]
- Syangtan, G.; Bista, S.; Dawadi, P.; Rayamajhee, B.; Shrestha, L.B.; Tuladhar, R.; Joshi, D.R. Asymptomatic SARS-CoV-2 Carriers: A Systematic Review and Meta-Analysis. Front. Public Health 2021, 8, 1066. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Laboratory Testing of 2019 Novel Coronavirus (2019-NCoV) in Suspected Human Cases: Interim Guidance, 17 January 2020. Available online: https://apps.who.int/iris/handle/10665/330676 (accessed on 16 November 2021).
- World Health Organization (WHO). Weekly Epidemiological Update on COVID-19—4 May 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---4-may-2021 (accessed on 16 November 2021).
- Serwis Rzeczypospolitej Polskiej. Pierwszy Przypadek Koronawirusa w Polsce—Ministerstwo Zdrowia—Portal Gov.Pl. Available online: https://www.gov.pl/web/zdrowie/pierwszy-przypadek-koronawirusa-w-polsce (accessed on 16 November 2021).
- Narodowy Instytut Zdrowia Publicznego PZH—PIB. COVID-19—Liczba Potwierdzonych Przypadków z Poszczególnych Powiatów Wg. Dane-i-Analizy.Pl—PZH. Available online: https://www.pzh.gov.pl/covid-19-liczba-potwierdzonych-przypadkow-z-poszczegolnych-powiatow-wg-dane-i-analizy-pl/ (accessed on 16 November 2021).
- Główny Urząd Statystyczny. Obszary Tematyczne/Ludność/Ludność. Available online: https://stat.gov.pl/obszary-tematyczne/ludnosc/ludnosc/ (accessed on 16 November 2021).
- Serwis Rzeczypospolitej Polskiej. Lista Laboratoriów COVID–Ministerstwo Zdrowia–Portal Gov.Pl. Available online: https://www.gov.pl/web/zdrowie/lista-laboratoriow-covid (accessed on 16 November 2021).
- Kowalska, M.; Hudzik, G.; Wodzisławska-Czapla, D.; Kaleta, A.; Zejda, J.E. Spatial Variability of SARS-CoV-2 Infection in the Silesian Voivodeship, Poland. Przegląd Epidemiol. 2020, 74, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Raciborski, F.; Pinkas, J.; Jankowski, M.; Sierpiński, R.; Zgliczyński, W.S.; Szumowski, Ł.; Rakocy, K.; Wierzba, W.; Gujski, M. Dynamics of the Coronavirus Disease 2019 Outbreak in Poland: An Epidemiological Analysis of the First 2 Months of the Epidemic. Pol. Arch. Intern. Med. 2020, 130, 615–621. [Google Scholar] [CrossRef]
- Paul, S.; Bhattacharya, S.; Mandal, B.; Haldar, S.; Mandal, S.; Kundu, S.; Biswas, A. Dynamics and Risk Assessment of SARS-CoV-2 in Urban Areas: A Geographical Assessment on Kolkata Municipal Corporation, India. Spat. Inf. Res. 2021, 29, 365–378. [Google Scholar] [CrossRef]
- Boterman, W.R. Urban-Rural Polarisation in Times of the Corona Outbreak? The Early Demographic and Geographic Patterns of the SARS-CoV-2 Epidemic in the Netherlands. Tijdschr. Voor Econ. Soc. Geogr. 2020, 111, 513–529. [Google Scholar] [CrossRef]
- Yang, S.; Stanzione, N.; Uslan, D.Z.; Garner, O.B.; de St Maurice, A. Clinical and Epidemiologic Evaluation of Inconclusive COVID-19 PCR Results Using a Quantitative Algorithm. Am. J. Clin. Pathol. 2021, 155, 376–380. [Google Scholar] [CrossRef]
- Engelmann, I.; Alidjinou, E.K.; Ogiez, J.; Pagneux, Q.; Miloudi, S.; Benhalima, I.; Ouafi, M.; Sane, F.; Hober, D.; Roussel, A.; et al. Preanalytical Issues and Cycle Threshold Values in SARS-CoV-2 Real-Time RT-PCR Testing: Should Test Results Include These? ACS Omega 2021, 6, 6528–6536. [Google Scholar] [CrossRef]
- Noh, J.Y.; Yoon, J.G.; Seong, H.; Choi, W.S.; Sohn, J.W.; Cheong, H.J.; Kim, W.J.; Song, J.Y. Asymptomatic Infection and Atypical Manifestations of COVID-19: Comparison of Viral Shedding Duration. J. Infect. 2020, 81, 816–846. [Google Scholar] [CrossRef]
- Xiao, A.T.; Tong, Y.X.; Gao, C.; Zhu, L.; Zhang, Y.J.; Zhang, S. Dynamic Profile of RT-PCR Findings from 301 COVID-19 Patients in Wuhan, China: A Descriptive Study. J. Clin. Virol. 2020, 127, 104346. [Google Scholar] [CrossRef]
- Sun, J.; Xiao, J.; Sun, R.; Tang, X.; Liang, C.; Lin, H.; Zeng, L.; Hu, J.; Yuan, R.; Zhou, P.; et al. Prolonged Persistence of SARS-CoV-2 RNA in Body Fluids. Emerg. Infect. Dis. 2020, 26, 1834–1838. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, L.; Liu, B.; Song, D. Case Report: Viral Shedding for 60 Days in a Woman with COVID-19. Am. J. Trop. Med. Hyg. 2020, 102, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hang, X.; Wei, B.; Li, D.; Chen, F.; Liu, W.; Yang, C.; Miao, X.; Han, L. Persistent SARS-COV-2 RNA Positivity in a Patient for 92 Days after Disease Onset: A Case Report. Medicine 2020, 99, e21865. [Google Scholar] [CrossRef]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV Viral Load Dynamics, Duration of Viral Shedding, and Infectiousness: A Systematic Review and Meta-Analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef]
- Yahav, D.; Yelin, D.; Eckerle, I.; Eberhardt, C.S.; Wang, J.; Cao, B.; Kaiser, L. Definitions for Coronavirus Disease 2019 Reinfection, Relapse and PCR Re-Positivity. Clin. Microbiol. Infect. 2021, 27, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Common Investigation Protocol for Investigating Suspected SARS-CoV-2 Reinfection. Available online: https://www.cdc.gov/coronavirus/2019-ncov/php/reinfection.html (accessed on 17 November 2021).
- Tillett, R.L.; Sevinsky, J.R.; Hartley, P.D.; Kerwin, H.; Crawford, N.; Gorzalski, A.; Laverdure, C.; Verma, S.C.; Rossetto, C.C.; Jackson, D.; et al. Genomic Evidence for Reinfection with SARS-CoV-2: A Case Study. Lancet Infect. Dis. 2021, 21, 52–58. [Google Scholar] [CrossRef]
- To, K.K.-W.; Hung, I.F.-N.; Ip, J.D.; Chu, A.W.-H.; Chan, W.-M.; Tam, A.R.; Fong, C.H.-Y.; Yuan, S.; Tsoi, H.-W.; Ng, A.C.-K.; et al. Coronavirus Disease 2019 (COVID-19) Re-Infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. 2021, 73, e2946–e2951. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.; Brodniak, S.L.; Voegtly, L.J.; Cer, R.Z.; Glang, L.A.; Malagon, F.J.; Long, K.A.; Potocki, R.; Smith, D.R.; Lanteri, C.; et al. A Case of Early Reinfection with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2021, 73, e2827–e2828. [Google Scholar] [CrossRef]
- Sevillano, G.; Ortega-Paredes, D.; Loaiza, K.; Zurita-Salinas, C.; Zurita, J. Evidence of SARS-CoV-2 Reinfection within the Same Clade in Ecuador: A Case Study. Int. J. Infect. Dis. 2021, 108, 53–56. [Google Scholar] [CrossRef] [PubMed]
- van Elslande, J.; Vermeersch, P.; Vandervoort, K.; Wawina-Bokalanga, T.; Vanmechelen, B.; Wollants, E.; Laenen, L.; André, E.; van Ranst, M.; Lagrou, K.; et al. Symptomatic Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection by a Phylogenetically Distinct Strain. Clin. Infect. Dis. 2021, 73, 354–356. [Google Scholar] [CrossRef]
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA 2020, 323, 1502–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Peng, J.; Xiong, Q.; Liu, Z.; Lin, H.; Tan, X.; Kang, M.; Yuan, R.; Zeng, L.; Zhou, P.; et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020, 59, 102960. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Kim, D.M.; Lee, H.; Ham, S.Y.; Oh, S.M.; Jeong, H.; Jung, J.; Kang, C.K.; Park, J.Y.; Kang, Y.M.; et al. Dynamics of Viral Load and Anti-SARS-CoV-2 Antibodies in Patients with Positive RT-PCR Results after Recovery from COVID-19. Korean J. Intern. Med. 2020, 36, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False Negative of RT-PCR and Prolonged Nucleic Acid Conversion in COVID-19: Rather than Recurrence. J. Med. Virol. 2020, 92, 1755–1756. [Google Scholar] [CrossRef] [Green Version]
- Trisnawati, I.; El Khair, R.; Puspitarani, D.A.; Fauzi, A.R.; Gunadi, G. Prolonged Nucleic Acid Conversion and False-Negative RT-PCR Results in Patients with COVID-19: A Case Series. Ann. Med. Surg. 2020, 59, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability Issues of RT-PCR Testing of SARS-CoV-2 for Hospitalized Patients Clinically Diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903–908. [Google Scholar] [CrossRef] [Green Version]
- Waked, R.; Makhoul, J.; Saliba, G.; Chehata, N.; Mortada, S.; Zoghbi, A.; Choucair, J.; Haddad, E. Are Two Consecutive Negative RT-PCR Results Enough to Rule out COVID-19? New Microbes New Infect. 2020, 37, 100750. [Google Scholar] [CrossRef]
- Feng, H.; Liu, Y.; Lv, M.; Zhong, J. A Case Report of COVID-19 with False Negative RT-PCR Test: Necessity of Chest CT. Jpn. J. Radiol. 2020, 38, 409–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arevalo-Rodriguez, I.; Buitrago-Garcia, D.; Simancas-Racines, D.; Zambrano-Achig, P.; del Campo, R.; Ciapponi, A.; Sued, O.; Martinez-García, L.; Rutjes, A.W.; Low, N.; et al. False-Negative Results of Initial RT-PCR Assays for COVID-19: A Systematic Review. PLoS ONE 2020, 15, e0242958. [Google Scholar] [CrossRef] [PubMed]
- Muenchhoff, M.; Mairhofer, H.; Nitschko, H.; Grzimek-Koschewa, N.; Hoffmann, D.; Berger, A.; Rabenau, H.; Widera, M.; Ackermann, N.; Konrad, R.; et al. Multicentre Comparison of Quantitative PCR-Based Assays to Detect SARS-CoV-2, Germany, March 2020. Eurosurveillance 2020, 25, 2001057. [Google Scholar] [CrossRef] [PubMed]
- Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020, 173, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Long, L.; Zhang, D.; Yuan, T.; Cui, S.; Yang, P.; Wang, Q.; Ren, S. Potential False-Negative Nucleic Acid Testing Results for Severe Acute Respiratory Syndrome Coronavirus 2 from Thermal Inactivation of Samples with Low Viral Loads. Clin. Chem. 2020, 66, 794–801. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, M.; Yuan, J.; Wang, F.; Wang, Z.; Li, J.; Zhang, M.; Xing, L.; Wei, J.; Peng, L.; et al. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection. Innovation 2020, 1, 100061. [Google Scholar] [CrossRef] [PubMed]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic Performance of Different Sampling Approaches for SARS-CoV-2 RT-PCR Testing: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef]
- Phan, T. Genetic Diversity and Evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81, 104260. [Google Scholar] [CrossRef]
- Peñarrubia, L.; Ruiz, M.; Porco, R.; Rao, S.N.; Juanola-Falgarona, M.; Manissero, D.; López-Fontanals, M.; Pareja, J. Multiple Assays in a Real-Time RT-PCR SARS-CoV-2 Panel Can Mitigate the Risk of Loss of Sensitivity by New Genomic Variants during the COVID-19 Outbreak. Int. J. Infect. Dis. 2020, 97, 225–229. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Detection of New SARS-CoV-2 Variants Related to Mink. 2020. Available online: https://www.ecdc.europa.eu/en/publications-data/detection-new-sars-cov-2-variants-mink (accessed on 19 August 2021).
- Brejová, B.; Boršová, K.; Hodorová, V.; Čabanová, V.; Reizigová, L.; Paul, E.D.; Čekan, P.; Klempa, B.; Nosek, J.; Vinař, T. A SARS-CoV-2 Mutant from B.1.258 Lineage with ∆H69/∆V70 Deletion in the Spike Protein Circulating in Central Europe in the Fall 2020. Virus Genes 2021, 57, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, X.; Ge, C.; Ding, Y.; Zhang, T.; Cao, S.; Meng, L.; Lu, S. Molecular Detection of SARS-CoV-2 Being Challenged by Virus Variation and Asymptomatic Infection. J. Pharm. Anal. 2021, 11, 257–264. [Google Scholar] [CrossRef] [PubMed]
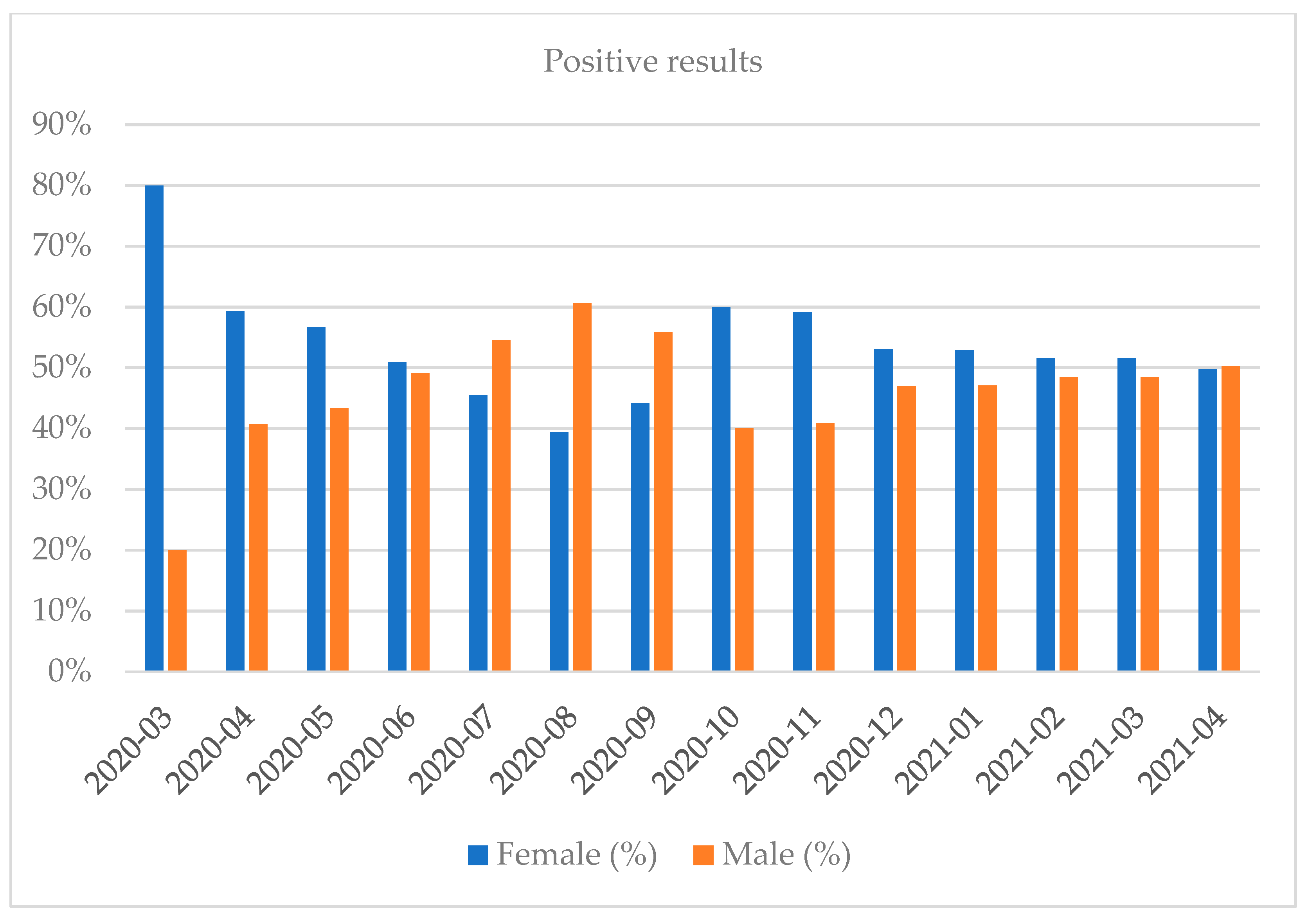
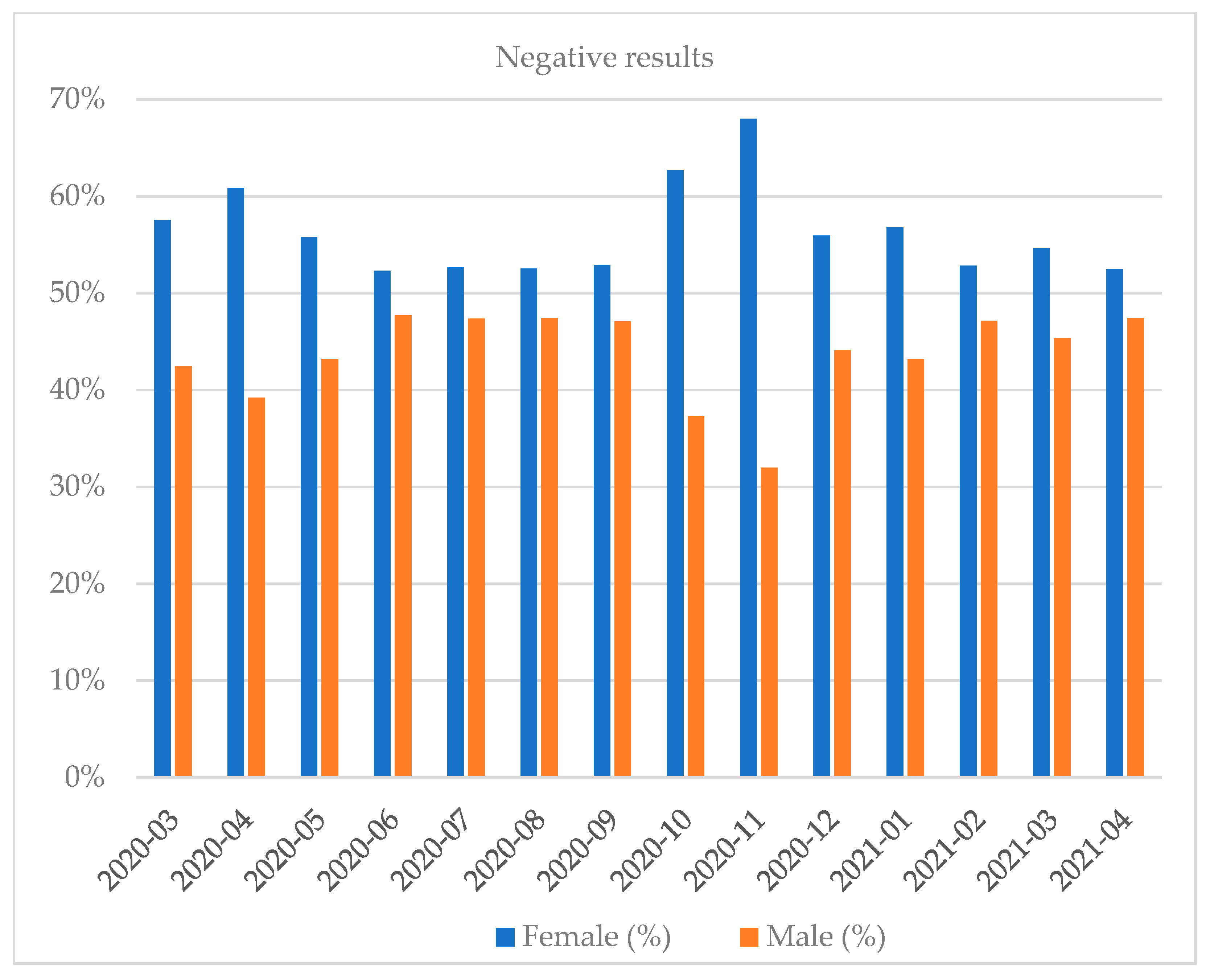
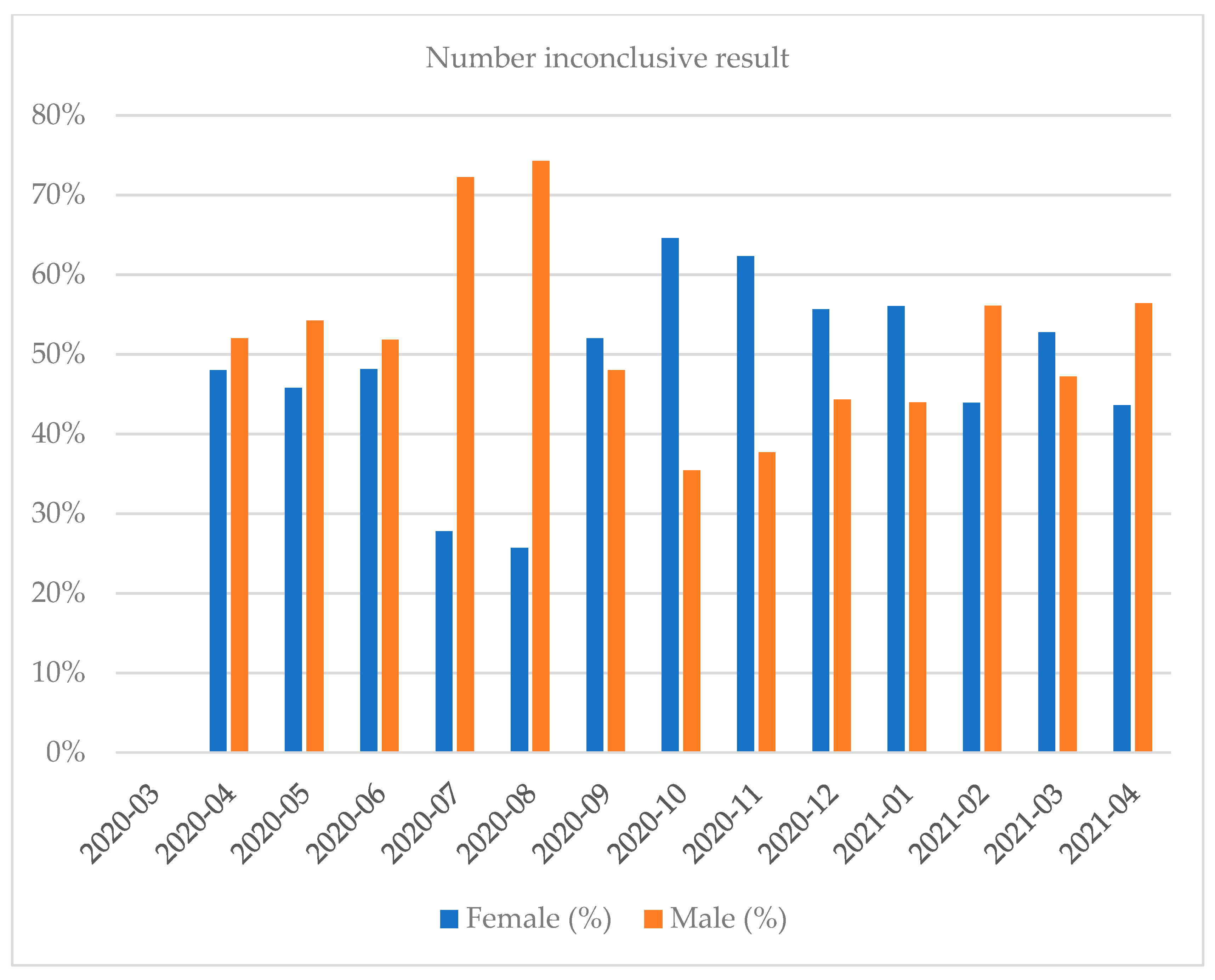
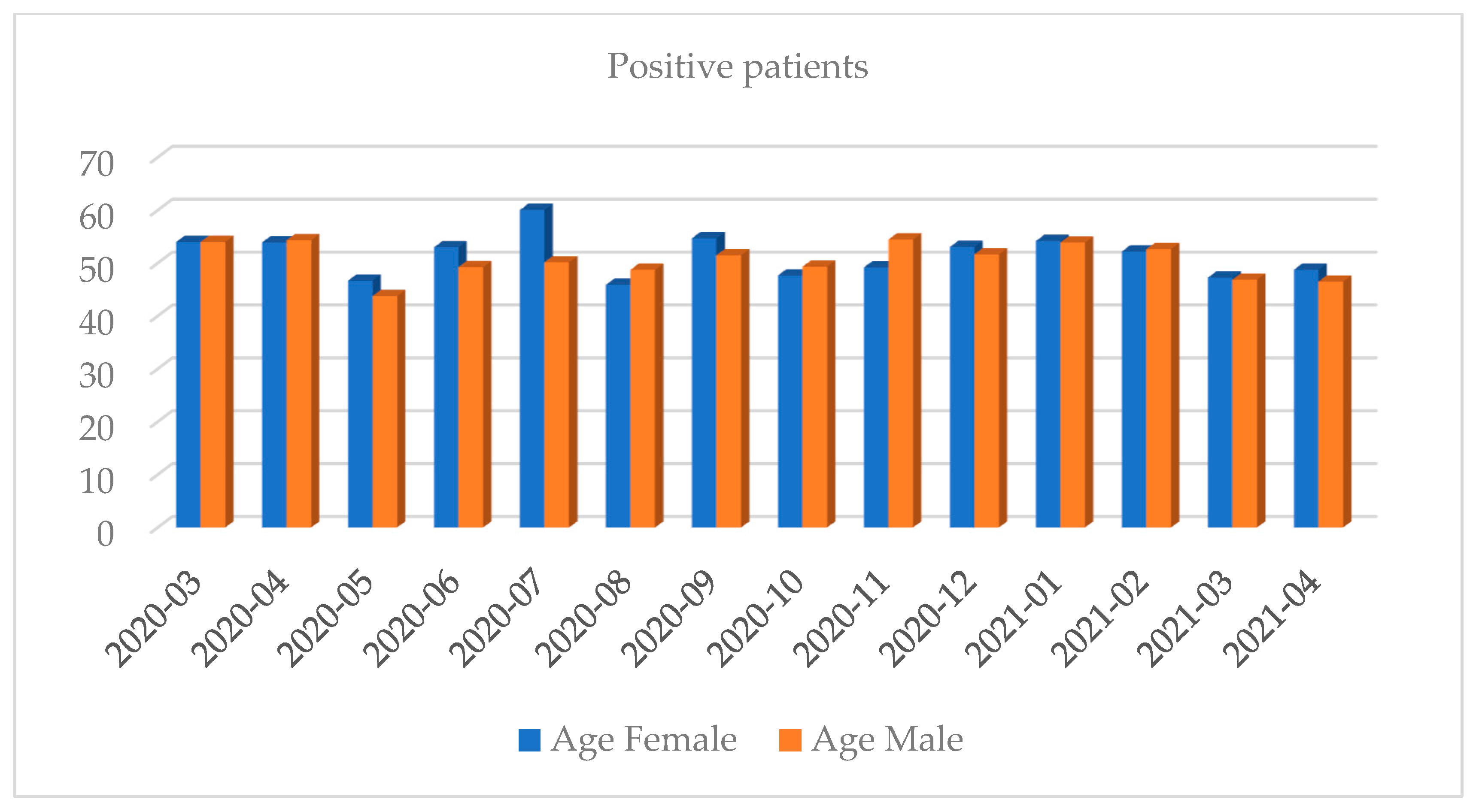
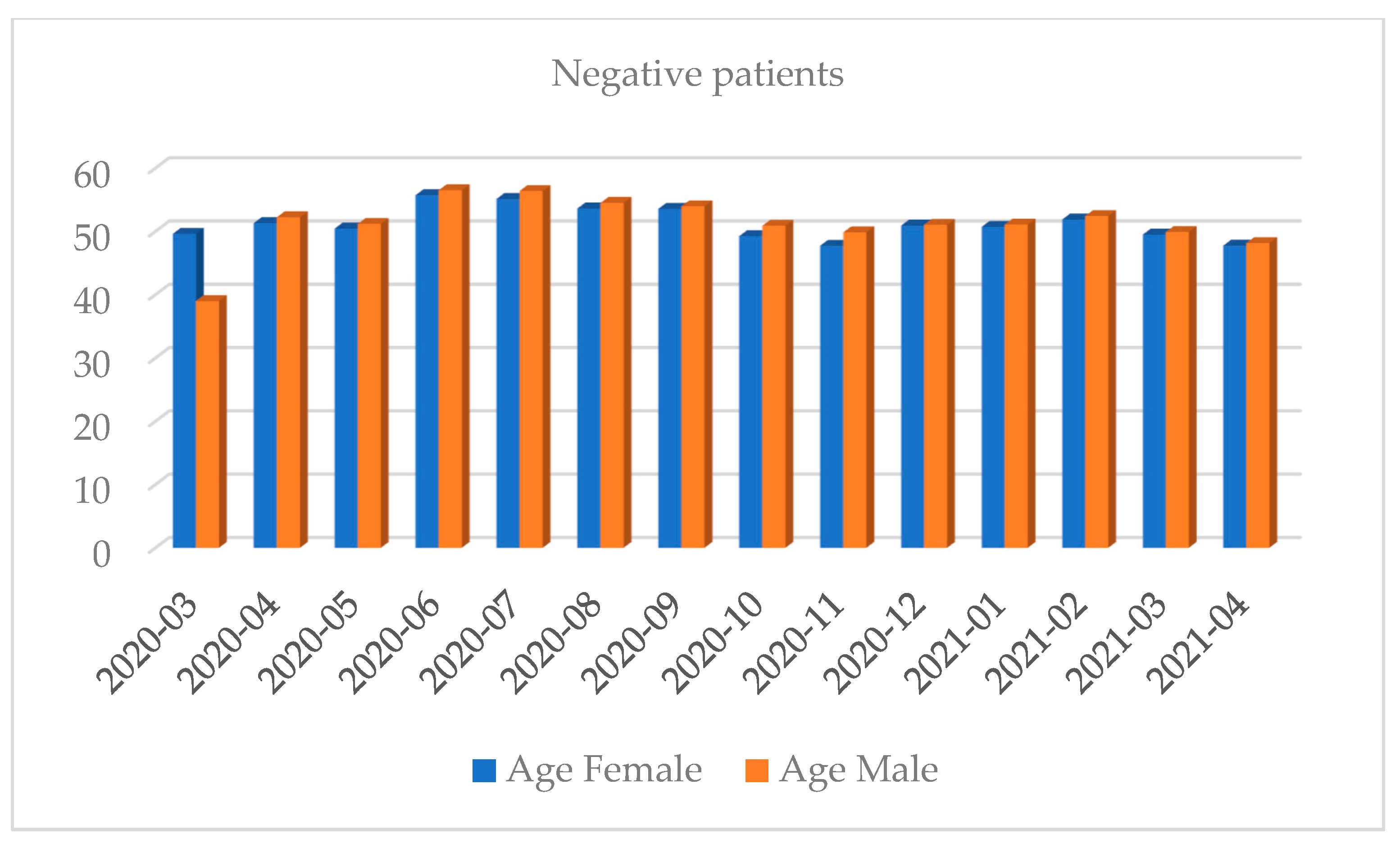
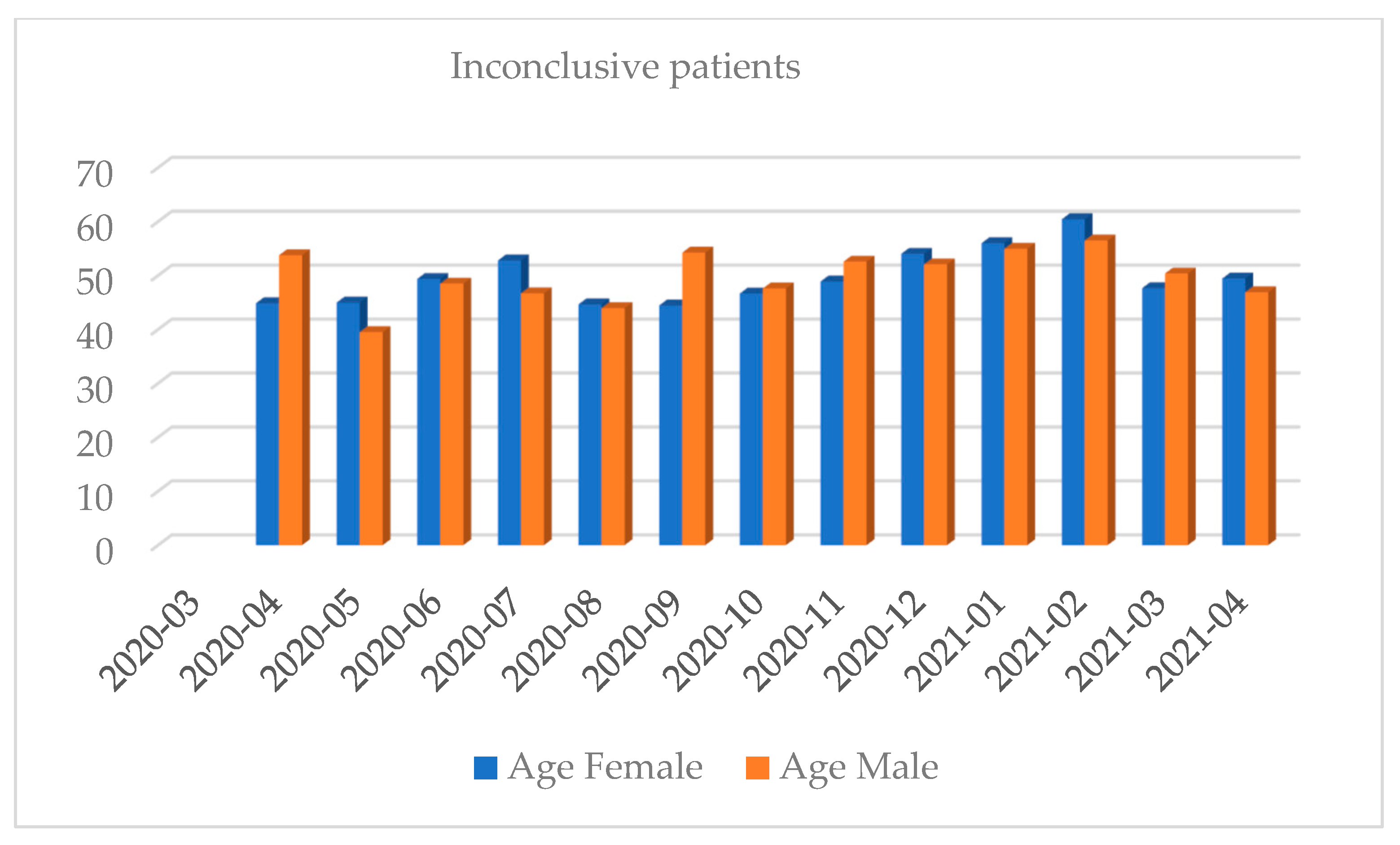
| Month and Year | Number of Tests | Positive Results, n (%) | Negative Results, n (%) | Number of Inconclusive Results, n (%) |
|---|---|---|---|---|
| 2020-03 | 144 | 5 (3.47) | 139 (96.53) | 0 (0.00) |
| 2020-04 | 4718 | 457 (9.69) | 4236 (89.78) | 25 (0.53) |
| 2020-05 | 9050 | 256 (2.83) | 8604 (95.07) | 190 (2.10) |
| 2020-06 | 8935 | 104 (1.16) | 8804 (98.53) | 27 (0.30) |
| 2020-07 | 6989 | 33 (0.47) | 6938 (99.27) | 18 (0.26) |
| 2020-08 | 7301 | 61 (0.84) | 7205 (98.69) | 35 (0.48) |
| 2020-09 | 6354 | 77 (1.21) | 6252 (98.39) | 25 (0.39) |
| 2020-10 | 13,792 | 3173 (23.01) | 10,266 (74.43) | 353 (2.56) |
| 2020-11 | 12,896 | 2676 (20.75) | 9936 (77.05) | 284 (2.20) |
| 2020-12 | 7324 | 1335 (18.23) | 5777 (78.88) | 212 (2.89) |
| 2021-01 | 7042 | 907 (12.88) | 5978 (84.89) | 157 (2.23) |
| 2021-02 | 6392 | 745 (11.66) | 5565 (87.06) | 82 (1.28) |
| 2021-03 | 10,411 | 2723 (26.16) | 7436 (71.42) | 252 (2.42) |
| 2021-04 | 7168 | 1788 (24.94) | 5208 (72.66) | 172 (2.40) |
| Total | 108,516 | 14,340 (13.21) | 92,344 (85.10) | 1832 (1.69) |
| Number of PCR Tests Performed | Number of Patients | Average Age (Years) | Female (%) | Minimal Interval between Two Extreme Positive Results (Days) | Average Interval between Two Extreme Positive Results (Days) | Maximum Interval between Two Extreme Positive Results (Days) |
|---|---|---|---|---|---|---|
| 2 | 362 | 58.42 ± 14.82 | 85.78 | 1.99 | 16.87 ± 9.99 | 52.88 |
| 3 | 46 | 67.22 ± 11.69 | 56.52 | 7 | 21.98 ± 4.79 | 37.67 |
| 4 | 9 | 72.22 ± 12.37 | 33.33 | 19.22 | 24.73 ± 3.73 | 33.09 |
| 5 | 3 | 79.67 ± 1.78 | 33.33 | 25.96 | 29.42 ± 2.59 | 33.31 |
| 6 | 1 | 83.00 ± ns | 100 | - | 35.41 ± ns | - |
| 8 | 1 | 81.00 ± ns | 100 | - | 48.71 ± ns | - |
| First Infection | Second Infection | |||
|---|---|---|---|---|
| Patients Number | GeneProof SARS-CoV-2 Vaccine Escape PCR Kit | VirellaSARS-CoV-2 Mutant Real Time RT-PCR Kit | GeneProof SARS-CoV-2 Vaccine Escape PCR Kit | VirellaSARS-CoV-2 Mutant Real Time RT-PCR Kit |
| 1. | NR | NR | Wildtype | Wildtype |
| 2. | Wildtype | Wildtype | NR | NR |
| 3. | Wildtype | Wildtype | NR | NR |
| 4. | Wildtype | B.1.258/Cluster V | UK | B.1.258/Cluster V |
| 5. | UK | B.1.258/Cluster V | Wildtype | NR |
| 6. | NR | NR | Wildtype | Wildtype |
| 7. | Wildtype | Wildtype | NR | NR |
| 8. | Wildtype | Wildtype | NR | NR |
| 9. | NR | NR | Wildtype | NR |
| 10. | Wildtype | B.1.258/Cluster V | NR | NR |
| 11. | Wildtype | Wildtype | Wildtype | Wildtype |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konka, A.; Lejawa, M.; Gaździcka, J.; Bochenek, A.; Fronczek, M.; Strzelczyk, J.K. RT-PCR Detection of SARS-CoV-2 among Individuals from the Upper Silesian Region—Analysis of 108,516 Tests. Diagnostics 2022, 12, 7. https://doi.org/10.3390/diagnostics12010007
Konka A, Lejawa M, Gaździcka J, Bochenek A, Fronczek M, Strzelczyk JK. RT-PCR Detection of SARS-CoV-2 among Individuals from the Upper Silesian Region—Analysis of 108,516 Tests. Diagnostics. 2022; 12(1):7. https://doi.org/10.3390/diagnostics12010007
Chicago/Turabian StyleKonka, Adam, Mateusz Lejawa, Jadwiga Gaździcka, Aneta Bochenek, Martyna Fronczek, and Joanna Katarzyna Strzelczyk. 2022. "RT-PCR Detection of SARS-CoV-2 among Individuals from the Upper Silesian Region—Analysis of 108,516 Tests" Diagnostics 12, no. 1: 7. https://doi.org/10.3390/diagnostics12010007
APA StyleKonka, A., Lejawa, M., Gaździcka, J., Bochenek, A., Fronczek, M., & Strzelczyk, J. K. (2022). RT-PCR Detection of SARS-CoV-2 among Individuals from the Upper Silesian Region—Analysis of 108,516 Tests. Diagnostics, 12(1), 7. https://doi.org/10.3390/diagnostics12010007







